The Hereditary Ontology for Phenoclinical Data
- This version:
- https://w3id.org/hereditary/ontology/phenoclinical/
- Latest version:
- https://w3id.org/hereditary/ontology/phenoclinical/
- Ontology Version IRI:
- https://w3id.org/hereditary/ontology/phenoclinical/1.0/
- Revision:
- 1.0
- Authors:
- Laura Menotti
- Gianmaria Silvello
- Download serialization:
-
- License:
- Cite as:
- Laura Menotti, Gianmaria Silvello. The Hereditary Ontology for Phenoclinical Data. Revision: 1.0. Retrieved from: https://w3id.org/hereditary/ontology/phenoclinical/schema/1.0/
Introduction back to ToC
The Clinical Data Modeling is based on the BrainTeaser Ontology (BTO) [Faggioli et al., 2024]. We followed the same event-based structure but added some properties and classes to include useful information to our use case. In particular, we added a class to represent the affiliated Socio-Health Local Authority (ULSS in Italy) for each patient, one for risk factors, and some properties for MS relapses. The biggest changes are in the Diagnostic Procedure area, where we included several new tests both for ALS and MS patients. Figure 1 reports the schema of the Clinical Data Modeling. The clinical course of ALS and MS patients is organized into eight semantic areas, each denoting a set of ontological classes: “Patient" with “Environmental Data", “Event", “Activity", “Contingencies", and “Intervention or Procedures", divided into “Surgical Procedures", “Diagnostic Procedures", and “Therapeutic Procedures".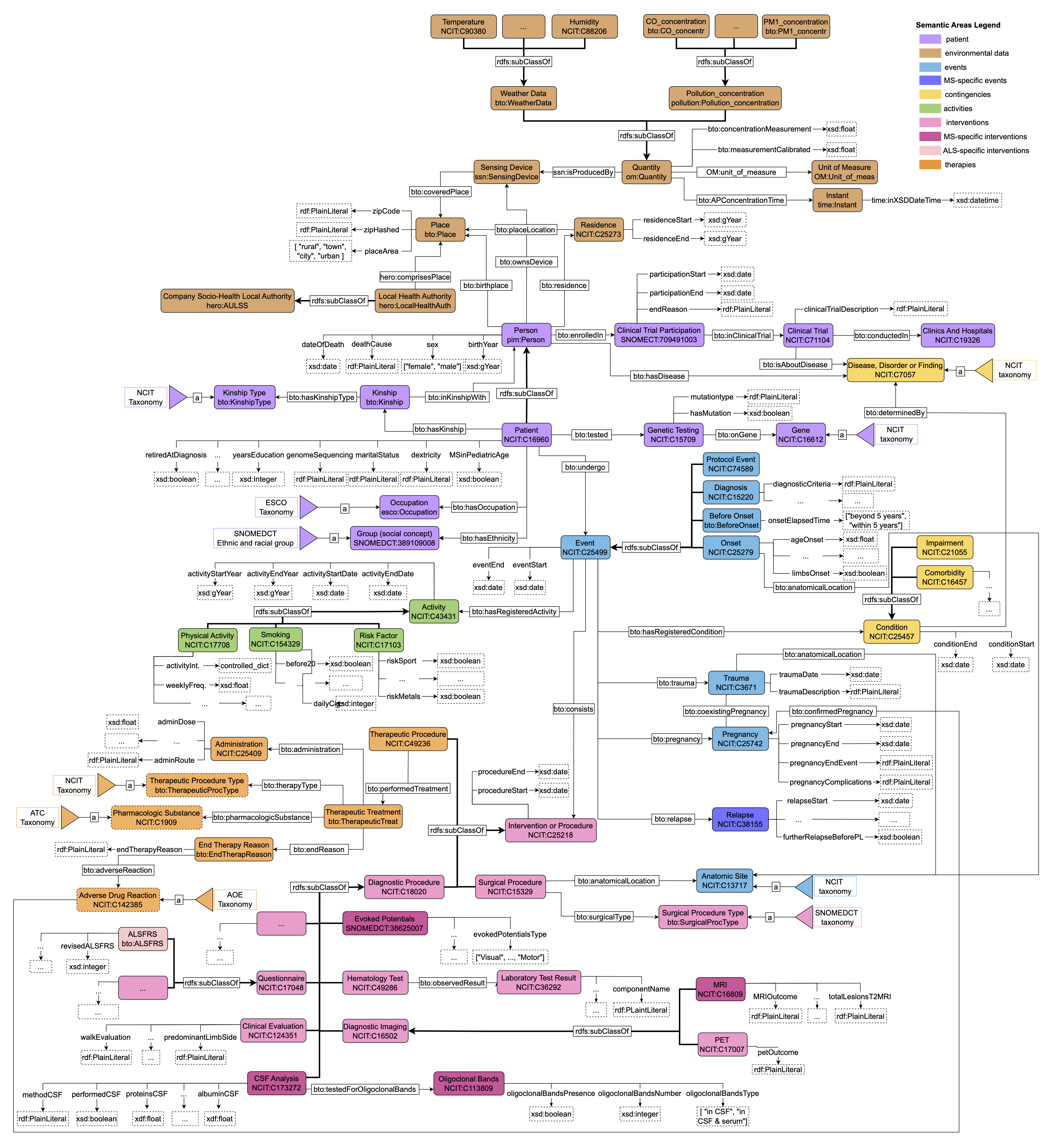
Namespace declarations
| Pollution | <http://ontology.eil.utoronto.ca/GCI/Environment/Pollution.owl#> |
| UATC | <http://purl.bioontology.org/ontology/UATC/> |
| [Ontology NS Prefix] | <https://w3id.org/hereditary/ontology/phenoclinical/schema/> |
| adms | <http://www.w3.org/ns/adms#> |
| contact | <http://www.w3.org/2000/10/swap/pim/contact#> |
| dc | <http://purl.org/dc/elements/1.1/> |
| foaf | <http://xmlns.com/foaf/0.1/> |
| isco | <http://data.europa.eu/esco/isco/> |
| ns | <http://creativecommons.org/ns#> |
| obo | <http://purl.obolibrary.org/obo/> |
| om-1 | <http://www.wurvoc.org/vocabularies/om-1.8/> |
| orcid | <https://orcid.org/0000-0002-0676-682> |
| owl | <http://www.w3.org/2002/07/owl#> |
| rdf | <http://www.w3.org/1999/02/22-rdf-syntax-ns#> |
| rdfs | <http://www.w3.org/2000/01/rdf-schema#> |
| schema | <https://w3id.org/brainteaser/ontology/schema/> |
| skos | <http://www.w3.org/2004/02/skos/core#> |
| ssn | <http://purl.oclc.org/NET/ssnx/ssn#> |
| terms | <http://purl.org/dc/terms/> |
| time | <http://www.w3.org/2006/time#> |
| vann | <http://purl.org/vocab/vann/> |
| xml | <http://www.w3.org/XML/1998/namespace> |
| xsd | <http://www.w3.org/2001/XMLSchema#> |
Overview back to ToC
This ontology has the following classes and properties.Classes
- Activity
- Administration
- Adverse Drug Reaction
- ALS Brainteaser Comorbidities
- Amyotrophic Lateral Sclerosis Functional Rating Scale (ALSFRS)
- Anatomic Site
- Beck Depression Inventory-II (BDI-II)
- Before Onset
- Blood Gas Analysis
- c6 h6 concentration
- Clinical Evaluation
- Clinical Trial
- Clinical Trial Participation
- Clinics and Hospitals
- CO concentration
- Cognitive Assessment
- Comorbidity
- Company-Socio-Health Local Autority
- Conditon
- Corpus Callosum Topography Score (CLTR)
- CSF Analysis
- Delis-Kaplan Executive Function System (D-KEFS)
- Delis-Kaplan Executive Function System Color-Word Interference Switching Condition (D-KEFS FSC)
- Delis-Kaplan Executive Function System Design Fluency Condition (D-KEFS FSD)
- Delis-Kaplan Executive Function System Sorting Test - Free Sort & Sort Recognition (D-KEFS SR)
- Diagnosis
- Diagnostic Imaging
- Diagnostic Procedure
- Disease, Disorder or Finding
- Electromyography
- End Therapy Reason
- Event
- Evoked Potentials
- Expanded Disability Status Scale (EDSS)
- Fatigue Severity Score (FSS)
- Gene
- Genetic Testing
- Group (social concept)
- Hematology Test
- Hoffman's Reflex Test
- Impairment
- Intervention or Procedure
- King's Clinical Staging Method
- Kinship
- KinshipType
- Laboratory Test Result
- Last 5 Years Event
- Lesion Topography Score (LTS)
- Lifestyle
- Local Health Autority
- Magnetic Resonance Imaging (MRI)
- Milano-Torino Functional Staging System (MiToS)
- More Than 5 Years Event
- Multiple Scleroris Neuropsychological Questionnaire (MSNQ)
- Muscle Biopsy
- Neurologic Exam
- Occupation
- Oligoclonal Bands
- Onset
- Paced Auditory Serial Addition Test (PASAT)
- Patient
- Person
- Pharmacologic Substance
- Physical Activity
- Place
- Plantar Reflex Test
- Positron Emission Tomography (PET)
- Pregnancy
- Protocol Event
- Pulmonary Function Test
- Questionnaire
- Relapse
- Residence
- Risk Factor
- Selective Reminding Test - Delayed
- Self-Reported Questionnaire
- Smoking
- Spatial Recall Test (SPART)
- Surgical Procedure
- Surgical Procedure Type
- Symbol Digit Modalities Test (SDMT)
- Symbol Digit Modalities Test - Delayed (SPART-D)
- Tendon Reflex Test
- Therapeutic Procedure
- Therapeutic Procedure Type
- Therapeutic Treatment
- Trauma
- Use-Case Comorbidities
- Word List Generation (WLG)
Object Properties
- administration
- adverseReaction
- anatomicalLocation
- APConcentrationTime
- birthplace
- coexistingPregnancy
- comprisesPlace
- conductedInClinic
- confirmedPregnancy
- consists
- coveredPlace
- determinedBy
- enrolledIn
- hasDisease
- hasEthnicity
- hasKinship
- hasKinshipType
- hasOccupation
- hasRegisteredActivity
- hasRegisteredCondition
- inClinicalTrial
- inKinshipWith
- is produced by
- isAboutDisease
- observedResult
- onGene
- performedTreatment
- pharmacologicSubstance
- placeLocation
- pregnancy
- relapse
- requiredMRI
- residence
- surgicalType
- tested
- testedForOligoclonalBands
- therapyType
- trauma
- undergo
- unit of measure
Data Properties
- abdominalSurgery
- acitivityStartYear
- activityEndYear
- activityType
- administrationDose
- administrationFrequency
- administrationRoute
- administrationUoM
- ageOnset
- albuminCSF
- alcoholUse
- alive
- alsfrs1
- alsfrs10
- alsfrs11
- alsfrs12
- alsfrs2
- alsfrs3
- alsfrs4
- alsfrs5
- alsfrs6
- alsfrs7
- alsfrs8
- alsfrs9
- alsfrsOriginal10
- alterationCSF
- ambulationEDSS
- autoimmuneDisease
- axialOnset
- before20Years
- betweenessCentralityCoefficient
- birthYear
- bloodGasPCO2
- bloodGasPH
- bloodGasPO2
- bloodGasSO2
- bodyMassIndex
- bowelAndBladderEDSS
- bowelAndBladderRelapse
- brainAtrophyMRI
- brainstemEDSS
- brainstemRelapse
- bulbarOnset
- bulbarSubscore
- cardiacDisease
- cerebellarEDSS
- cerebellarRelapse
- cerebralEDSS
- cervicalSpineSurgery
- cervicalTrauma
- clinicallyEvaluatedEDSS
- clinicalTrialDescription
- clusteringCoefficient
- cognitiveRelapse
- comorbiditySeverity
- comorbidityTreatment
- componentName
- concentrationMeasurement
- conditionEnd
- conditionStart
- conductionBlocks
- contiguousLesionsMRI
- dailyCigarettes
- deathCause
- deathDate
- dextricity
- diabetes
- diagnosticCriteria
- diet
- dyslipidemia
- educationLevel
- electricTraumaRisk
- electromyographyOutcome
- emotionalLiability
- endReason
- endTherapyReason
- eventEnd
- eventStart
- everSmoked
- evokedPotentialsLocation
- evokedPotentialsResponse
- evokedPotentialsType
- formatDKEFS
- furtherRelapseBeforePL
- gadoliniumLesionsT1MRI
- gadoliniumLesionsT1MRINumber
- generalizedOnset
- genomeSequencing
- globalCoefficient
- glucoseCSF
- granulocytesCSF
- hadMajorTrauma
- hadSurgicalIntervention
- hasMutation
- headNeckSurgery
- headTrauma
- height
- herbicidesRisk
- hoffmanLeft
- hoffmanRight
- hypertension
- IgGIndexCSF
- immunoglobulinGCSF
- intensity
- kingsValue
- leftPlantarReflex
- lesionsT1MRI
- leukocytesCountCSF
- limbsOnset
- lowerLimbsTendonReflex
- lowerLimbSurgery
- lumboSacralSpineSurgery
- lumboSacralTrauma
- lymphocytesCSF
- maritalStatus
- massetericReflex
- measuredLevel
- measurementCalibrated
- menopauseStatus
- metalsRisk
- methodCSF
- mitosValue
- moreThan10PercentWeightloss
- motorSubscore
- msDiagnosis2010Criteria
- MSInPaediatricAge
- multipleSclerosisType
- muscleAtrophy
- muscleBiopsyOutcome
- muscleTone
- mutationType
- neckTrauma
- newOrEnlargedLesionsT2MRI
- newOrEnlargedLesionsT2MRINumber
- normalLevel
- notes
- oligoclonalBandsNumber
- oligoclonalBandsPresence
- oligoclonalBandsType
- onsetElapsedTime
- originalALSFRS
- otherOnset
- otherRelapse
- packYear
- participationEnd
- participationStart
- pathologicalJCCSF
- pelvicSurgery
- performedCSF
- performedCSFAnalysis
- performedMuscleBiopsy
- pesticidesRisk
- petOutcome
- placeArea
- polioRisk
- positivityResult
- predominantLimbsImpairment
- predominantLimbsSide
- predominantMotorNeuron
- pregnancyComplications
- pregnancyEnd
- pregnancyEndEvent
- pregnancyStart
- primaryNeoplasm
- procedureEnd
- procedureStart
- proteinsCSF
- pulmonaryAOPAbs
- pulmonaryFEV6
- pulmonaryFVCRel
- pulmonaryMEPAbs
- pulmonaryMIPAbs
- pulmonaryPCO2
- pulmonaryPO2
- pulmonaryVCAbs
- pyramidalEDSS
- pyramidalRelapse
- Questionnaire's score reported by a patient.
- quotientAlbuminCSF
- raoClassification
- relapseClinicalTherapy
- relapseCortisoneTherapy
- relapseDuration
- relapseEnd
- relapseHospitalAdmission
- relapseImpactOnADL
- relapseRecovery
- relapseSequela
- relapseSeverity
- relapseStart
- residenceEnd
- residenceStart
- respiratorySubscore
- retiredAtDiagnosis
- revisedALSFRS
- rightPlantarReflex
- sensoryEDSS
- sensoryRelapse
- sex
- sexuality
- solventsRisk
- sportRisk
- stroke
- sunExposure
- thoracicSpineSurgery
- thoracicSurgery
- thoracicTrauma
- thyroidDisorder
- tongueAtrophy
- tongueFasciculation
- tongueNotProtruding
- tongueProtrusion
- tongueThrust
- totalLesionsT2MRI
- traumaDate
- traumaDescription
- traumaRisk
- unitOfMeasure
- upperLimbsMobility
- upperLimbsTendonReflex
- upperLimbSurgery
- urinaryUrgency
- urinaryUrgencyTherapy
- virusJCTestedCSF
- visualEDSS
- visualEvokedPotentialAmplitude
- visualEvokedPotentialLatency
- visualRelapse
- walkEvaluation
- weeklyFrequency
- weight
- zipCode
- zipHashed
Named Individuals
- Abdomen
- Abdominal Pain
- Abdominoplasty
- ACE Inhibitors, Plain
- Acetylcarnitine
- Acetylsalicylic Acid, Combinations excl. Psycholeptics
- Aciclovir
- Acquired Hypothyroidism
- Acute Cystitis
- Acute Myocardial Infarction
- Acute Pyelonephritis
- Acute Respiratory Failure
- Acute Tonsillitis
- Ademetionine
- Adenocarcinoma
- Administration of Ineffective Drug
- Administrative and commercial managers
- Adverse Drug Event
- Adverse Drug Reaction
- Agents Acting on the Renin-Angiotensin System
- Ageusia
- Agricultural, forestry and fishery labourers
- Alanine Aminotransferase Increased
- Alcohol Dependance
- Alectinib
- Alemtuzumab
- Alimentary Tract and Metabolism
- All Other Non-Therapeutic Products
- All Other Therapeutic Products
- Allergens
- Allergic Asthma
- Allergic Rhinitis
- Alpha-Lipoic Acid
- Alprazolam
- Alzheimer's Disease
- Amantadine
- Amenorrhea
- Amenorrhea due to Pituitary Dysfunction
- Amiodarone
- Amitriptyline
- Amitriptyline and Psycholeptics
- Amlodipine
- Amoxicillin and Beta-Lactamase Inhibitor
- Amyotrophic Lateral Sclerosis
- Anabolic Agents for Systemic Use
- Analgesics
- Anatomic Site
- Anemia
- Anesthetics
- Angioedema
- Ankle Fracture
- Ann Arbor Stage IV Mantle Cell Lymphoma
- Ann Arbor Stage IV T-Cell Non-Hodgkin Lymphoma
- Anorectal Fistula
- Anorexia
- Anosmia
- Anthelmintics
- Anti-Acne Preparations
- Anti-Parkinson Drugs
- Antianemic Preparations
- Antibacterials for Systemic Use
- Antibiotics and Chemotherapeutics for Dermatological Use
- Anticonvulsant Agent
- Antidiarrheals, Intestinal Antiinflammatory/Antiinfective Agents
- Antiemetics and Antinauseants
- Antiepileptics
- Antifungals for Dermatological Use
- Antigout Preparations
- Antihemorrhagics
- Antihistamines for Systemic Use
- Antihypertensives
- Antiinfectives for Systemic Use
- Antiinflammatory and Antirheumatic Products
- Antimycobacterials
- Antimycotics for Systemic Use
- Antineoplastic Agents
- Antineoplastic and Immunomodulating Agents
- Antiobesity Preparations, excl. Diet Products
- Antiparasitic Products, Insecticides and Repellents
- Antiprotozoals
- Antipruritics, incl. Antihistamines, Anesthetics, etc.
- Antipsoriatics
- Antispetic and Disinfectants
- Antithrombotic Agents
- Antivirals for Systemic Use
- Anxiety Disorder
- Appendectomy
- Appetite Stimulants
- AR Gene
- Armed forces occupations
- Armed forces occupations, other ranks
- Arrhythmia
- Arthritis
- Asian (ethnic group)
- Aspiration Pneumonitis
- Assemblers
- Asthma
- Atenolol
- Atenolol and Other Diuretics
- Atopic Dermatitis
- Atrial Fibrillation
- Atrial Flutter
- Atrovastatin
- Aunt
- Autoimmune Disease
- Autoimmune Nervous System Disorder
- Autoimmune Pancytopenia
- Azathioprine
- Baclofen
- Bacterial Pneumonia
- Baker Cyst
- Barbexaclone
- Barnidipine
- Basal Cell Carcinoma
- BCG Vaccine
- Behavioral Impairment
- Bell's Palsy
- Beta Blocking Agents
- Betahistine
- Betamethasone
- BG-12
- Bile and Liver Therapy Drugs
- Bile Therapy
- Biotin
- Bipolar Disorder
- Bisoprolol
- Black (ethnic group)
- Black African
- Blepharitits
- Blepharospasm
- Blood and Blood Forming Organs
- Blood Substitutes and Perfusion Solutions
- Bone Marrow
- Botulinum Toxin
- Bowel Obstruction
- Bradycardia
- Brain
- Brain Stem
- Breast Carcinoma
- Bromazepam
- Brother
- Brotizolam
- Brown-Sequard Syndrome
- Building and related trades workers, excluding electricians
- Business and administration associate professionals
- Business and administration professionals
- C9orf72 Gene
- Cabergoline
- Calcifediol
- Calcium Channel Blockers
- Calcium Folinate
- Calcium Homeostasis
- Calcium, Combinations with Vitamin D and/or Other Drugs
- Cannabinoids
- Carbamazepine
- Carcinoma
- Cardiac Therapy
- Cardiovascular Disorder
- Cardiovascular System
- Cardioversion
- Caucasian
- Cavernous Hemangioma
- Ceftriaxone
- Celiac Disease
- Central Nervous System Degenerative Disorder
- Cerebellum
- Cervical Spinal Cord
- Cervical Spine
- Cetirizine
- Chalazion
- Chicken Pox
- Chief executives, senior officials and legislators
- Chlorphenamine
- Cholecystectomy
- Chronic Atropic Gastritis
- Chronic Lung Disorder
- Cinnarizine
- Cinnarizine, Combinations
- Citalopram
- Cladribine
- Cladribine
- Cleaners and helpers
- Cleft Palate
- Clerical support workers
- Clobazam
- Clonazepam
- Clopidogrel
- Closed Dislocation of Shoulder
- Closed Fracture of Rib
- Closed Fracture of Tibia
- Codeine, Combinations excl. Psycholeptics
- Cognitive Impariment
- Cold Sore
- Colecalciferol
- Colon Adenocarcinoma
- Commissioned armed forces officers
- Common Bile Duct Stone
- Cone Biopsy
- Congenital Adrenal Hyperplasia
- Contrast Media
- Coronary Artery Bypass Graft
- Corticosteroids for Systemic Use
- Corticosteroids, Dermatological Preparations
- Corticotropin
- Cortisone Therapy for Relapse
- Cough and Cold Preparations
- Cousin
- COVID-19 Infection
- Covid-19 Vaccines
- Craft and related trades workers
- Cranial Nerve VII Palsy
- Customer services clerks
- Cutaneous Melanoma
- Cyanocobalamin
- Cyclophosphamide
- Cyst
- Daclizumab
- Dantrolene
- Daughter
- DCTN1 Gene
- Deep Vein Thrombosis
- Deflazacort
- Dementia
- Denosumab
- Depression
- Dermatologicals
- Desogestrel and Ethinylestradiol
- Dexamethasone
- Diabetes Mellitus
- Diabetic Ketoacidosis
- Diagnostic Agents
- Diagnostic Radiopharmaceuticals
- Diazepam
- Dietary Supplement
- Digestives, incl. Enzymes
- Dimethyl Fumarate
- Diphtheria Vaccines
- Disease or Disorder
- Disease, Disorder or Finding Taxonomy
- Diuretics
- Dog Bite
- Domperidone
- Drainage of Bartholin's Abscess
- Drivers and mobile plant operators
- Drospirenone and Ethinylestradiol
- Drug Hypersensitivity AE
- Drug Hypersensitivity Syndrome
- Drug Intolerance AE
- Drugs for Acid Related Disorders
- Drugs for Bile Therapy and Lipotropics in Combination
- Drugs for Constipation
- Drugs for Functional Gastrointestinal Disorders
- Drugs for MS Symptomatic Therapy
- Drugs for MS Therapy Side Effects
- Drugs for Obstructive Airway Diseases
- Drugs for Other Diseases
- Drugs for Treatment of Bone Diseases
- Drugs Used in Diabetes
- Dyslipidemia
- Ectoparasiticides, incl. Scabicides, Insecticides and Repellents
- Ecunerv
- Eczema
- Electrical and electronic trades workers
- Elementary occupations
- Emergency Tracheostomy
- Emollients and Protectives
- Enalapril
- Endocrine Therapy
- Endometriosis
- Epidermal Inclusion Cyst
- Escitalopram
- Esomeprazole
- Essential Hypertension
- Ethnic Group
- Etizolam
- Etoricoxib
- Excision of Basal Cell Carcinoma
- Excision of Benign Neoplasm
- Excision of Cyst
- Excision of dermatofibroma
- Excision of Neoplasm
- Extrapyramidal Disorder
- Eye
- Fampridine
- Father
- Favism
- Femur Fracture
- Fever Chills
- Fibroma
- Fibromyalgia
- Finding
- Fingolimod
- First Cousin
- First Degree Relative
- Flu-Like Symptoms
- Flunarizine
- Fluoxetine
- Flurazepam
- Food preparation assistants
- Food processing, wood working, garment and other craft and related trades workers
- Frontotemporal Dementia
- Fungal Infection
- Furosemide
- FUS Gene
- Gabapentin
- Gastritis
- Gastroenteritits
- Gastroesophageal Reflux
- Gastroplasty
- Gastryc Bypass
- Gene
- General and keyboard clerks
- General Nutrients
- Generalized Epilepsy
- Genito Urinary System and Sex Hormones
- Gianmaria Silvello
- Giorgio Maria Di Nunzio
- Glatiramer Acetate
- Glaucoma
- Gliclazide
- Glimepiride
- Glucocorticoids, Systemic
- Granddaughter
- Grandfather
- Grandmother
- Grandson
- Group Taxonomy
- Guglielmo Faggioli
- Gynecological Antiinfectives and Antiseptics
- Handicraft and printing workers
- Hashimoto Thyroiditis
- Head
- Head and Neck Approaches
- Head and Neck Finding
- Head and Neck Surgery
- Head Trauma
- Headache
- Health associate professionals
- Health professionals
- Heart Failure
- Hepatic Toxicity
- Hepatitis
- Hepatitis A Infection
- Hepatitis B, Purified Antigen
- Hereditary Factor XI Deficiency
- Hereditary Hemolytic Anemia
- Hernia
- Herpes Simplex Keratitis
- Herpes Simplex Virus Infection
- Herpes Zoster
- High Grade Cervical Squamous Intraepithelial Neoplasia
- Hispanic
- Hospital General Universitario Gregorio Marañón
- Hospitality, retail and other services managers
- Hydrocortisone
- Hydrocortisone
- Hydroxychloroquine
- Hypercholesterolemia
- Hypersensitivity
- Hypertension
- Hypertensive Crisis
- Hyperthyroidism
- Hypnotics and Sedatives
- Hypothyroidism
- Hysterectomy
- Ibuprofen
- Ichthyosis
- Immune Sera and Immunoglobulins
- Immunoactive Drugs
- Immunoglobulins, Normal Human, for Intravascular Adm.
- Immunostimulants
- Immunosuppressants
- Indometacin
- Infectious Mononucleosis
- Information and communications technicians
- Information and communications technology professionals
- Inguinal Hernia
- Injury
- Insertion of Implantable Intrathecal Access System
- Institute of Molecular Medicine Joao Lobo Antunes
- Insulin Glargine
- Insulin Lispro
- Insulins and Analogues
- Interferon Beta Natural
- Interferon Beta-1a
- Interferon Beta-1b
- Interstitial Pneumonia
- Intervertebral Disc Prolapse
- Intestinal Pseudo-Obstruction
- Iridocyclitis
- Iron-Deficiency Anemia
- Keratitis
- Keratoconus
- Ketorolac
- Kinship Type Taxonomy
- Klippel-Feil Syndrome
- Labourers in mining, construction, manufacturing and transport
- Lactulose
- Lamotrigine
- Langerhans Cell Histiocytosis
- Laura Menotti
- Legal, social and cultural professionals
- Legal, social, cultural and related associate professionals
- Lercanidipine
- Leukopenia
- Levetiracetam
- Levodopa and Decarboxylase Inhibitor
- Levothyroxine Sodium
- Limb
- Lipid Modifying Agents
- Liver Therapy, Lipotropics
- Long QT Syndrome
- Losartan
- Lower Extremity
- Lumbar Hernia
- Lumbar Puncture Headache
- Lumbar Spine
- Lumbosacral Region
- Lymphopenia
- Malgradate
- Malignant Colon Neoplasm
- Malignant Neoplasm
- Malignant Skin Neoplasm
- Malignant Thyroid Gland Neoplasm
- Managers
- Market-oriented skilled agricultural workers
- Market-oriented skilled forestry, fishery and hunting workers
- Maternal Aunt
- Maternal Uncle
- Medicated Dressings
- Melanocytic Nevus
- Melatonin
- Mesalazine
- Metal, machinery and related trades workers
- Metformin
- Methotrexate
- Methotrexate
- Methylprednisolone
- Migraine
- Migraine With Aura
- Mineral Supplements
- Mitoxantrone
- Mitrazapine
- Modafinil
- Molluscum Contagiosum Virus
- Mondino Foundation
- Monoclonal B_Cell Lymphocytosis
- Monoclonal Gammopathy
- Mood Disorder
- Mother
- Motor Neuron Disease
- Multiple Sclerosis
- Mumps
- Muscle Relaxants
- Musculo-Skeletal System
- Myasthenia Gravis
- Myeloproliferative Neoplasm
- Naltrexone
- Nasal Preparations
- Natalizumab
- Nebivolol
- Neck
- Neck Trauma
- Neonatal Hearing Impairment
- Neoplasm
- Nephew
- Nervous System
- Neurofibromatosis
- Neutropenic Disorder
- Nicergoline
- Nicola Ferro
- Niece
- Non-commissioned armed forces officers
- Non-Invasive Mechanical Ventilation (NIV)
- Non-Neoplastic Central Nervous System Disorder
- Numerical and material recording clerks
- Obesity
- Obstructive Sleep Apnea Syndrome
- Occupation Taxonomy
- Ocrelizumab
- Olanzapine
- Olmesartan Medoxomil
- Operation on Abdominal Region
- Operative Procedure on Lumbosacral Spinal Structure
- Operative Procedure on Thoracic Spinal Structure
- Operative Procedure on Upper Extremity
- Ophthalmologicals
- Ophthalmologicals and Otologicals Products
- Opioid Use Disorder
- Optic Neuritis
- OPTN Gene
- Osteoporosis
- Other Alimentary Tract and Metabolism
- Other clerical support workers
- Other Dermatological Preparations
- Other Drugs for Disorders of the Musculo-Skeletal System
- Other Gynecologicals
- Other Hematological Agents
- Other Nervous System Drugs
- Other Respiratory System Products
- Other Vaccines
- Otologicals
- Oxcarbazepine
- Oxybutynin
- Paliperidone
- Palmar Fibromatosis
- Pancreatic Hormones
- Pancytopenia
- Pantoprazole
- Paracetamol
- Parkinson's Disease
- Paroxetine
- Partial Cystectomy
- Paternal Aunt
- Paternal Uncle
- Peginterferon Alfa-2b
- Peginterferon Beta-1a
- Pelvis
- Penicillin Allergy
- Percutaneous Endoscopic Gastrostomy (PEG)
- Perindopril and Amlodipine
- Peripheral Vasodilators
- Personal care workers
- Personal service workers
- Personality Disorder
- Pharmacologic Substance
- Phenobarbital
- Phenytoin
- Pituitary and Hypothalamic Hormones and Analogues
- Pituitary Neuroendocrine Tumor
- Plant and machine operators and assemblers
- Plasmapheresis
- Pleural Effusion
- Pneumococcal Pneumonia
- Pneumonia
- Pneumonia Caused By SARS-CoV-2
- Poliomyelitis, Trivalent, Inactivated, Whole Virus
- Post-Herpetic Neuralgia
- Pramipexole
- Prasugrel
- Prazepam
- Prednisolone
- Prednisone
- Pregabalin
- Preparations for Treatment of Wounds and Ulcers
- Primary Neoplasm
- Primary Progressive Aphasia
- Procedure on Pelvic Region of Trunk
- Production and specialised services managers
- Professionals
- Progestogens and Estrogens Systemic Contraceptives, Sequential Preparations
- Proglumide
- Progressive Multifocal Leukoencephalopathy
- Prolapsed Lumbar Intervertebral Disc
- Propanolol
- Protective services workers
- Proteinuria
- Psoriasis
- Psoriatic Arthritis
- Psychiatric Disorder
- Psychoanaleptics
- Psycholeptics
- Pulmonary Embolism
- Pulmonary Sarcoidosis
- Pulmonary Thromboembolism
- Quetiapine
- Radius Fracture
- Ramipril
- Reboxetine
- Refuse workers and other elementary workers
- Relative
- Removal of Acoustic Neuroma
- Renal Colic
- Repair of Bladder
- Respiratory System
- Retinal Detachment
- Rheumatoid Arthritis
- Rifampicin
- Riluzole
- Rituximab
- Rivaroxaban
- Rivastigmine
- Ropinirole
- Sales workers
- Sarcoidosis
- Scarlet Fever
- Science and engineering associate professionals
- Science and engineering professionals
- Second Degree Relative
- Secukinumab
- Seizure Disorder
- Sensory Organs
- Serositis
- Sertraline
- Serum Glutamic Pyruvic Transaminase, CTCAE
- Service and sales workers
- Sex Hormones and Modulators of the Genital System
- Shoulder Dislocation
- Sign
- Sign or Symptom
- Sildenafil
- Simvastatin
- Sinusitis
- Siponimod
- Sister
- Sitagliptin
- Sjogren Syndrome
- Skilled agricultural, forestry and fishery workers
- Skin Basal Cell Carcinoma
- Skin Basosquamous Cell Carcinoma
- Skin Carcinoma
- Skin Cyst
- SOD1 Gene
- Son
- Sphincter
- Spina Bifida
- Spinal and Bulbar Muscular Atrophy, X-linked 1
- Spinal Cord
- SQSTM1 Gene
- Staphylococcal Infection
- Stationary plant and machine operators
- Stefano Marchesin
- Stomatological Preparations
- Street and related sales and service workers
- Stroke
- Subdural Hematoma
- Subsistence farmers, fishers, hunters and gatherers
- Sulfasalazine
- Supratentorial Brain
- Surgical Dressings
- Surgical Procedure on Cervical Spine
- Surgical Procedure on Lower Extremity
- Surgical Procedure on Thorax
- Surgical Procedure Type Taxonomy
- Symptom
- Syncope
- Systemic Hormonal Preparations, excl. Sex Hormones and Insulins
- Tamoxifen
- Tamsulosin
- TARDBP Gene
- TBK1 Gene
- Teaching professionals
- Technicians and associate professionals
- Telmisartan
- Teriflunomide
- Tetanus Immunoglobulin
- Tetracosactide
- Thalassemia
- Therapeutic Procedure Type Taxonomy
- Therapeutic Radiopharmaceuticals
- Thiamazole
- Thiethylperazine
- Third Degree Relative
- Thoracic Spinal Cord
- Thoracic Spine
- Thoracic Trauma
- Thorax
- Throat Preparations
- Thrombocytopenia
- Thromboembolism
- Thyroid Gland Disorder
- Thyroid Gland Excision
- Thyroid Therapy
- Thyroiditis
- Tibia Fracture
- Ticlopidine
- Tinea Versicolor
- Tizanidine
- Tocilizumab
- Tolterodine
- Tonics
- Tooth Abscess
- Topical Products for Joint and Muscular Pain
- Topiramate
- Toxoplasmosis
- Transient Ischemic Attack
- Transurethral Prostatectomy
- Trastuzumab
- Trazodone
- Triazolam
- Trigeminal Nerve Disorder
- Trospium
- Two Vessel Coronary Disease
- Type 1 Diabtes Mellitus
- Type 2 Diabetes Mellitus
- Uncle
- University of Padua
- University of Turin
- Upper Extremity
- Ureteral Stenosis
- Urologicals
- Ursodeoxycholic Acid
- Ustekinumab
- Uterine Corpus Leiomyoma
- Vaccines
- Valaciclovir
- Valproic Acid
- Various
- Vasoprotectives
- Venlafaxine
- Verapamil
- Vertebral Column
- Vertigo
- Vigabatrin
- Viral Pericarditis
- Vitamins
- Vitiligo
- Vulvovaginal Candidiasis
- Vulvovaginitis
- Warfarin
- Zolmitriptan
- Zolpidem
- Zuclopenthixol
Methodology back to ToC
Design Principles
In the following, we report how HERO has been designed to meet the Open Biological and Biomedical Ontology Foundry (OBO) and FAIR principles also favoring its adoption in heterogeneous scenarios. The detailed description of each of the OBO principle is available at https://obofoundry.org/principles/fp-000-summary. html.- First, the ontology is Open and publicly available. It is possible to find its definition and description at http://hereditary.dei.unipd.it/ontology/phenoclincal/.
- Secondly, the ontology schema is defined according to the OWL 1.2 Common Format
- The proposed ontology relies on a unique URI/Identifier Space identified by the prefix https://w3id.org/hereditary/ontology/phenoclincial/schema/.
- A description of the Versioning procedure is available as part of the documentation of HERO.
- The Scope of HERO is clearly defined: the ontology is meant to model the anamnestic and clinical history of patients affected by two neurological diseases, ALS and MS.
- Following the OBO principles, we associate Textual Definitions to each class of the ontology, to also favour its re-usage in other scenarios.
- Before defining a new relation, Relations available on the Relations Ontology (RO) have been accurately considered. None of the HERO relations presents the same meaning and could have been replaced with one of the RO – nevertheless, this possibility has been always attentively scrutinized.
- A detailed Documentation of the ontology is available on its web page.
- For what concerns Documented Plurality of Users and Commitment To Collaboration, these aspects are intrinsic in the development and usage of the HERO ontology. In fact, HERO has been developed in the context of the HEREDITARY Project, which includes partners from multiple European countries. The co-design approach used to devise HERO defines its collaborative nature.
- HERO clearly identifies its Locus of Authority into its developers, who are indicated on the web page of the ontology.
- HERO follows strict Naming Conventions described below.
- Finally, the HEREDITARY consortium is actively working on the Maintenance and update of HERO.
Co-design Approach
To design the HERO, we adopted a co-design approach, strictly collaborating with the medical partners and domain experts, in order to embed their knowledge in the HERO and, at the same time, to validate all the design choices. To this end, we operated in an iterative way, producing several intermediate versions of the ontology, and discussing them with our domain experts. We exploited the iterative process of discussion with the medical partners to make sure that these newly defined concepts correctly described the corresponding real-world concepts and to guarantee the semantic quality of the ontology.Implementation Principles
To provide consistency in HERO we expressed some basic principles to adopt when defining classes and properties. These guidelines involve external referencing, anno- tation properties, and naming conventions.External Referencing Whenever we model clinical information, it is preferable to use external ontologies to provide correspondence between largely used medical terminology. Due to its widely diffused adoption and exhaustiveness, our primary taxonomy of choice is NCIT [30], but we also employed other well-known resources when no information is available in NCIT, e.g. SNOMED-CT or ATC. In general, this is common practice when developing ontologies [44]. In fact, by reusing entities and properties already defined in other ontologies, we are not only able to enforce collaboration and data consistency among databases but to guarantee the authori- tativeness of the semantic meaning of these resources. External referencing has been managed with annotation properties and using the URI of the term in the original taxonomy. In general, we used external URIs when we defined named individuals that refer to abstract concepts. On the contrary, when we inserted a new class in HERO we used the Hereditary namespace and connected references using annotation properties.
Namespaces HERO URIs are divided into two namespaces: the schema namespace https://w3id.org/hereditary/ontology/schema/ and the resource namespace https://w3id.org/hereditary/ontology/resource/. All URIs corresponding to classes, data properties, and object properties belong to the former namespace. At the same time, in the latter, we include all URIs referring to real-world instances of the entities described in HERO at an ontological level. Notice that, in this sense, the resource namespace is empty until the practitioner starts populating it with real-world data. The only instances included in the schema namespace are the named individuals corresponding to Simple Knowledge Organization System (SKOS) concepts (as defined in Subsection 3.4). Our choice of including these elements in the schema namespace stems from the fact that, akin to relational modeling controlled dictionaries, these entities do not depend on the data underneath but can be seen as a predefined thesaurus of concepts and are a fundamental part of the environment that we model.
Classes Definition and Annotation Properties All components of the HERO have additional information in the form of annotation properties. We defined a list of essential metadata to add when we introduced a new class. Firstly, all classes must have a label denoting the name and a comment, which provides a brief explana- tion together with its source. If the class had an equivalent in NCIT, we inherited the name and definition. In this case, we also added another annotation property called “rdfs:isDefinedBy with the Internationalized Resource Identifier (IRI) corre- sponding to the NCIT term of reference. Most biomedical vocabularies are mapped in the UMLS with a unique identifier called Concept Unique Identifier (CUI) [40]. For each class that had a UMLS reference, we added the annotation property “dcterms:conformsTo” with the URL of the corresponding concept.
Naming Conventions Regarding object and data properties, we applied a similar approach and defined the needed annotations and naming conventions to provide uniformity throughout HERO. In particular, all components must have a label and a comment. About object properties, we used explanatory labels where we included the property range. In this case, the comment explains the relationship between the two classes involved. Concerning data properties, the label usually includes the name of the domain class so that its meaning is intuitive. We also added a comment with the attribute description and, when available, the definition source. In conclusion, we defined all data properties to achieve consistency when specifying datatypes. For example, all string properties have datatype “rdf:PlainLiteral”. We also used the “note” annotation property in all HERO components for additional remarks or business logic rules.
Usage of the Simple Knowledge Organization System (SKOS)
In ontology design, it is good practice to use external taxonomies [44] – which are not necessarily ontologies – to ensure correspondence with the most common terminology, especially in the medical domain. This is common practice when developing ontologies, in fact by reusing entities and properties already defined in other ontologies, we are not only able to enforce the collaboration and data consistency among database, but to guarantee the authoritativeness of the semantic meaning of these resources. In HERO, external resources have been employed for example to model the diseases affecting a patient, anatomical sites of traumas, and pharmacological substances. The common aspect between these elements is that we are interested in the abstract concept behind the medical term. To avoid classism – the design pattern where an external hierarchy is modeled as a hierarchy of ontological classes – whenever we modeled a classification scheme that used an external taxonomy we employed the SKOS data model. Each component of the referenced ontology has been modeled as a named individual that belongs to two classes, SKOS Concept and the one in HERO. This has two important advantages: i) it allows to save space, by dramatically reducing the number of needed URI ii) it reduces the complexity of the queries. The disadvantage of this approach is that it prevents us from describing the peculiarities of the specific head of the specific patient. Nevertheless, since it is not a requirement of our ontology, we opted for the modeling pattern described above. We employed such a design principle for each class that refers to a set of abstract terms and does not have any data or object property associated. All classes modeled using the SKOS standard and the corresponding taxonomy of reference are reported in the table below.| Class | Reference Taxonomy |
|---|---|
| Kinship Type | National Cancer Institute Thesaurus (NCIT) |
| Occupation | Occupations pillar of the ESCO Classification (ESCO) |
| Group (social concept) | SNOMED Clinical Terms (SNOMED-CT) |
| Gene | National Cancer Institute Thesaurus (NCIT) |
| Disease, Disorder or Findings | National Cancer Institute Thesaurus (NCIT) |
| Surgical Procedure Type | SNOMED Clinical Terms (SNOMED-CT) |
| Therapeutic Procedure Type | National Cancer Institute Thesaurus (NCIT) |
| Anatomic Site | National Cancer Institute Thesaurus (NCIT) |
| Pharmacological Substance | Anatomical Therapeutic Chemical (ATC) Classification |
| Advesre Drug Reaction | Ontology of Adverse Events (OAE) |
Description back to ToC
The Hereditary ontology models clinical and genomics data to provide a unifying layer to query different database sources through an Ontology-Based Data Access (OBDA) framework. This part of the ontology focuses on phenoclinical data for ALS and MS patients. . The Clinical Data Modeling is based on the BrainTeaser Ontology (BTO) [Faggioli et al., 2024]. We followed the same event-based structure but added some properties and classes to include useful information to our use case. In particular, we added a class to represent the affiliated Socio-Health Local Authority (ULSS in Italy) for each patient, one for risk factors, and some properties for MS relapses. The biggest changes are in the Diagnostic Procedure area, where we included several new tests both for ALS and MS patients.Patient Modeling
BTO is a tool to model each person’s disease progression, with the objective of helping understand its progression. To better contextualize the patient’s clinical history, several pieces of information need to be recorded. In particular, BTO focuses on data about each patient’s lifestyle, clinical events, and clinical family history. Such information is modeled in the “Patient” semantic area, as illustrated in Figure 2.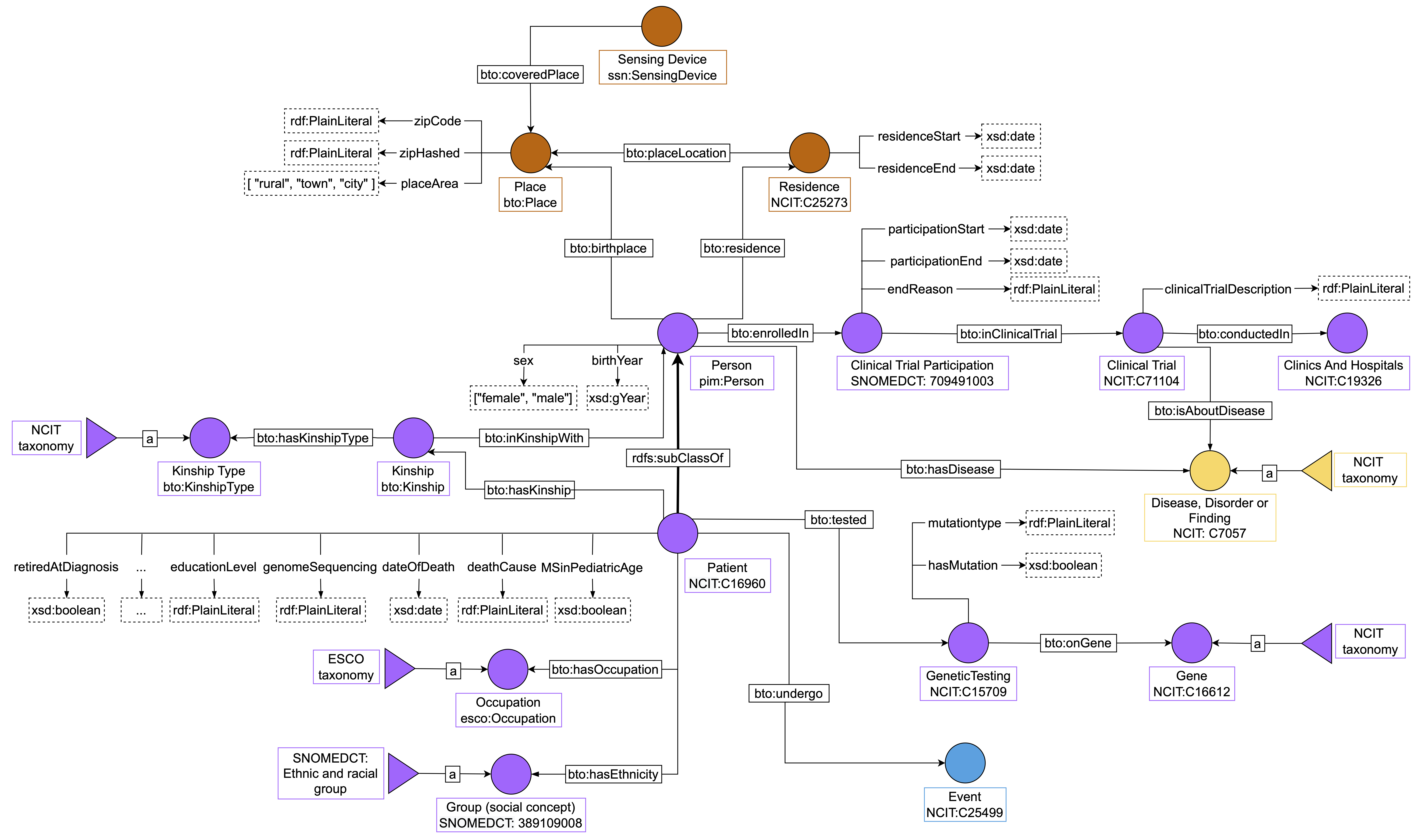
Figure 2: Patient semantic area. “Patient” is a subclass of the class Person from FOAF ontology. We directly connect to patient data about genetic mutations, occupation, family history, and residence place. The latter is useful to link environmental information to each patient, which is modeled by importing the “Global City Indicator Environment Ontology” [38].
Environmental Data
Besides genetic predisposition, lifestyle and environmental factors are shown to
affect the risk of MS. Once we acknowledge such correlation, some of these environmental
and lifestyle factors can be modified with prevention measures, especially
for people at greater risk [45]. On the other hand, identifying environmental factors
linked to ALS has proven to be challenging and it still lacks large cohorts. In both
cases, understanding the interaction between genetic and environmental factors can
be a great resource for integrating precision medicine into ALS or MS care [46].
That is why connecting environmental data to each patient in BTO is a fundamental
requirement. These environmental data include pollution information for a given
location, recorded by a weather station. To store this piece of information, in BTO
the practitioner can record each patient’s birth and residence places using the class
“Place”. To model environmental data, we imported Global City Indicator Environment
Ontology[38], also known as Pollution ontology, an ontology designed to
represent ontologically environmental data registered by weather stations. In par-
ticular, we connected a weather station, modeled with the class “Sensing Device”
from the above-mentioned ontology, to a location, modeled with the class “Place”,
with the object property “:coveredPlace”, to model the fact that a station registers
environmental data for a given location. We report how we integrated air pollutants in
Figure 3. The concentration of each air pollutant of interest is
modeled as a subclass of “Air Pollution Concentration”. Notice that we are interested in
storing information about Particulate Matter (PM) 10, PM 2.5, Ozone (O3), Nitrogen
Dioxide (NO2), Sulphur Dioxide (SO2), and Carbon Monoxide (CO) thus we
created a class in our namespace for pollutants not represented in the imported
ontology, such as CO. Air pollution concentrations are produced by sensing de-
vices, thus we connected the classes “Air Pollution Concentration” and “Sensing
Device” with the property “ssn:isProducedBy”. The concentration value of
each air pollutant is stored using the data property “numerical value”, while
the date of the measure is referred to with the object property ":APConcentrationTIme" that links
“Air Pollution Concentration” class to class “time:Instant”. All classes, object
properties, and data properties mentioned above are imported from the “Global
City Indicator Environment Ontology”[38].
Figure 3: Environmental data integration using the imported “Global City Indicator
Environment Ontology” [38]. We linked the class “Place” with “Sensing
Device”, which produces pollution concentration data. Such information
is stored using one of the subclasses of “Air Pollution Concentration”, which
represents the specific pollutant.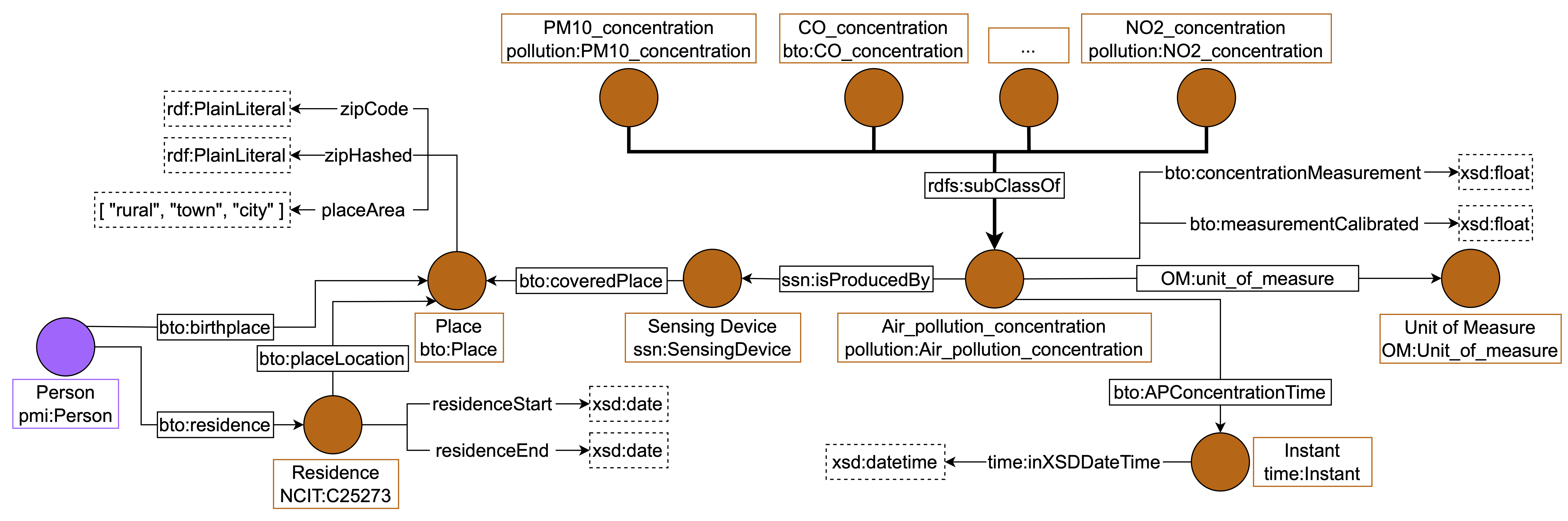
Event Modeling
The clinical history of a patient is not directly connected to the “Patient” class itself, but we assume such data to be registered during a clinical visit where, for instance, the physician fills a form with patient information regarding any relevant clinical event, physical activities, habits, or any arisen clinical condition communicated by the patient to the clinician during the visit. For this reason, we modeled the “Event” semantic area as follows. We specified four subclasses of the class “Event”, each with its specific data prop- erties. The possible event types are: i) the “Before Onset” event, describing as a cumulative event, everything that happened before the onset of the symptoms and for which there are no specific temporal details; ii) the “Onset”, describing the event when a patient experimented the symptoms for the first time; iii) the Diagnosis event, describing the event and exams carried out when the patient was diagnosed with the disease; iv) a “Protocol Event”, representing any possible occurrence happened after the diagnosis. A visual depiction of the modeling approach is reported in Figure 4.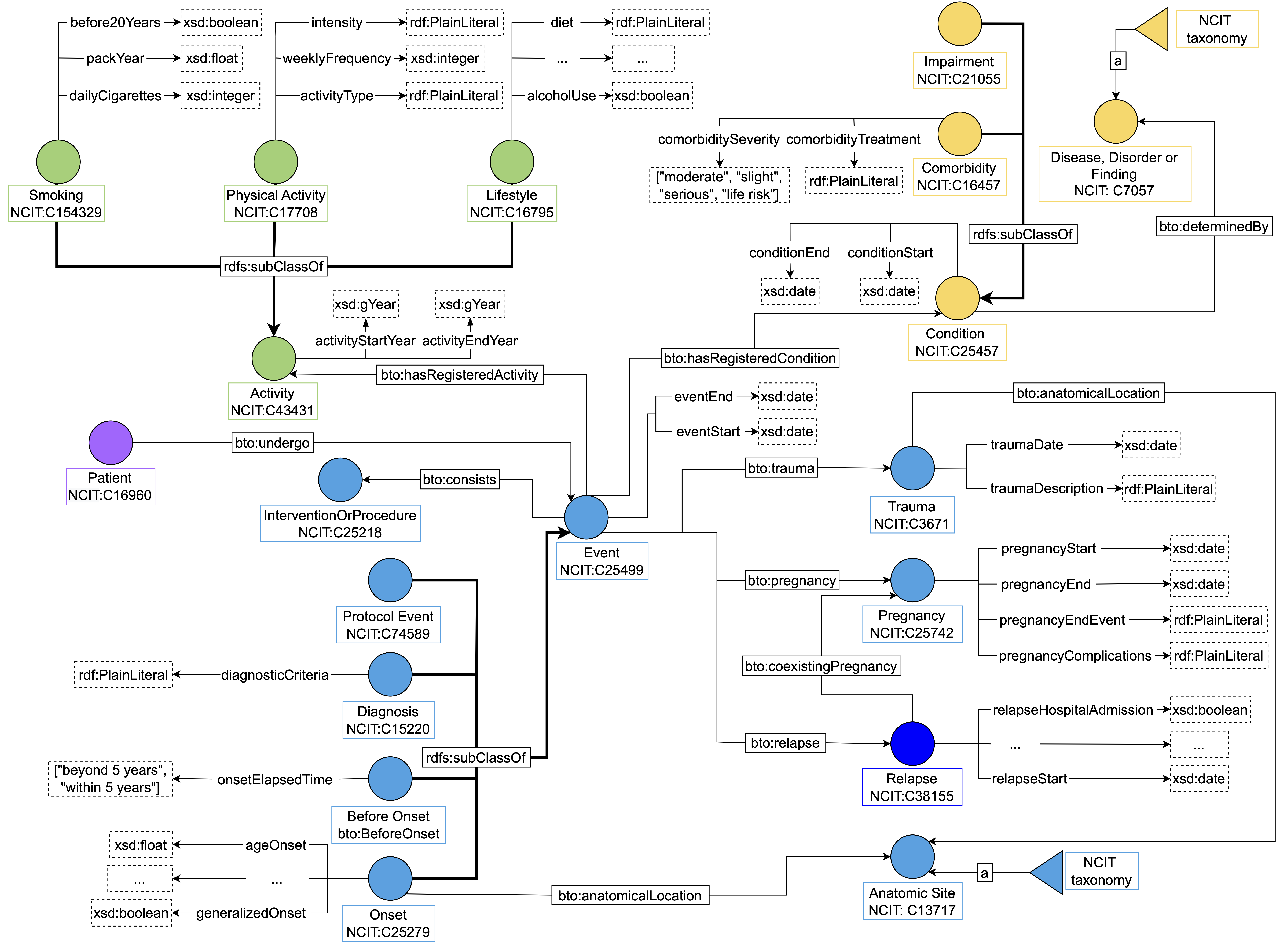
Figure 4: Event semantic area. Each patient is connected to clinical history and lifestyle information through the class Event. In fact, using such nodes physicians can store data about habits, past traumas, any clinical event (class “Condition”), coexisting medical conditions (called “comorbidities”), or pregnancies and MS patients’ relapses.
Disease, Disorder or Finding Taxonomy
The class “Disease, Disorder or Finding” includes diseases, like carcinoma or chickenpox infection, but also injuries, symptoms, and findings. This allows modeling and storing in a KB any sort of clinical event that occurred to a patient, even if it is not directly linked to ALS or MS. We modeled this class following the SKOS data model, using NCIT as reference taxonomy, with a few additions from SNOMED-CT thesaurus. As illustrated in Figure 4, we modeled past traumas with the class “Trauma”. However, some injuries have also been made available in the “Disease, Disorder or Finding” taxonomy. As a general rule, we use the taxonomy whenever the trauma is specific (e.g., “Open Dislocation of the Shoulder”). On the other hand, if the clinician knows only that a patient suffered from generic trauma to the shoulder, we instantiate an individual of type “Trauma” and connect it to its Anatomic Site (e.g., “shoulder”) with the property “:anatomicalLocation”. Notice that, thanks to SKOS characteristics, specific traumas (e.g., “open dislocation of the shoulder”) are in relation “:hasBroaderTransitive” with the general concept “injury”. We follow a similar line of thought to deal with symptoms. In this case, we use the taxonomy to model cases where the practitioner needs to store information about general symptoms (e.g., headache, fever). A particular group of symptoms that required deeper modeling granularity is the one that constitutes the onset of the disease (either ALS or MS). Given the high relevance that such symptoms have on the course of the disease, they have not been modeled using the symptoms taxonomy but as data properties of the class “Onset”.
Intervention Modeling
According to the proposed modeling paradigm, an event might include one or more interventions – for example, a visit might include multiple exams and multiple pre- scriptions of therapeutic substances. Therefore the “Intervention or Procedures” semantic area comprises surgical procedures, diagnostic tests, and therapeutic treat- ments. Figure 5 illustrates the main classes and properties involved in this semantic area. “Intervention or procedure” has three subclasses – “Surgical Procedure”, “Diagnostic Procedure” and “Therapeutic Procedure” – each denoting different operations.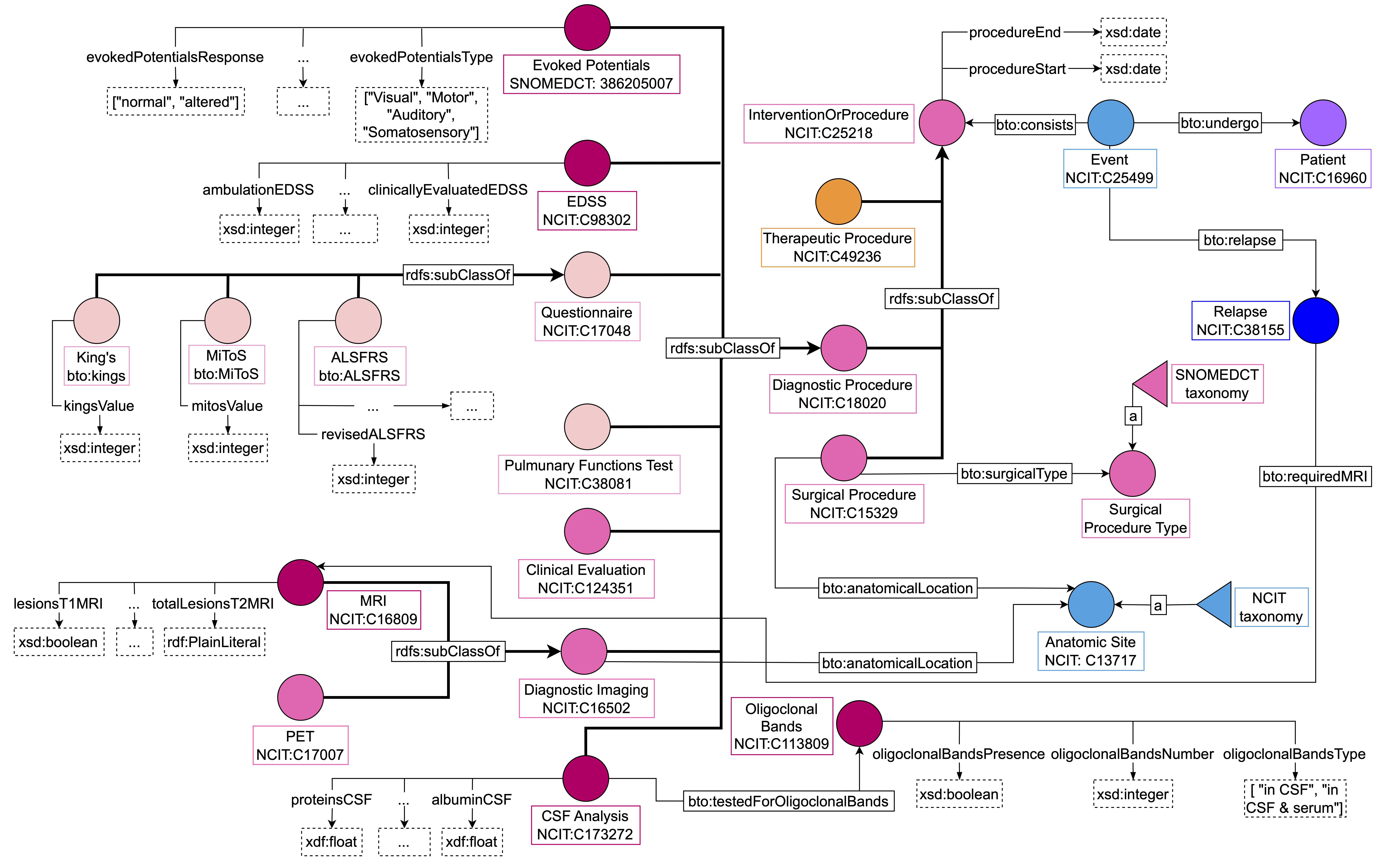
Figure 5: “Intervention or Procedures” area. We distinguish between surgical procedures, diagnostic procedures, and therapeutic procedures. Both surgical and therapeutic interventions are the same for MS and ALS patients while diagnostic procedures can differ based on the disease.
"Surgical Procedures” includes all treatments that involve surgery. Since different surgery procedures might vary widely, in BTO we define the class “Surgery Procedure Type” to represent different surgery types. This class is modeled following the SKOS standard and using the SNOMED-CT taxonomy as a reference. Furthermore, we linked the “Surgical Procedure” class to “Anatomic Site” to allow clinicians to store information about which body region was affected by the surgery. In this way, one can both instantiate the surgery type or the anatomical location based on the information available or the need of the developed application.
Diagnostic procedures differ based on MS or ALS patients. For instance, MS patients can take an Evoked Potentials test or a CSF analysis and their level of disability can be monitored over time using the EDSS. On the contrary, ALS patients are tested on their pulmonary function and the disease progression can be assessed using the Revised Amyotrophic Lateral Sclerosis Functional Rating Scale (ALSFRS-R), the King’s clinical staging method (KINGS), or the Milano-Torino functional staging system (MiToS). Diagnostic intervention information can be a great reason for assessing the disease progression of each patient. For example, with BTO we can return the number of patients having an EDSS score in between some ranges, which can be an indicator of impairment severity in patients affected by MS. One peculiarity is that hematology test components have been modeled as a class called “Laboratory Test Result” where the clinician can specify the component name, the measured level, its unit of measure, and whether the result was normal. We also included a data property called “:positivyResult” to express the presence or absence of specific blood components (e.g., antibodies).
Figure 6 reports the diagnostic procedure semantic area modeled in HERO. We highlighted with different colors the novel components added to HERO compared to BTO. We added some properties to classes “CSF Analysis" and “Clinical Evaluation" to include walking status assessments and additional CSF method information. Concerning ALS patients, we added six new classes, two representing neurological exam and blood gas analysis, while the others represent the assessment of muscle response to nerve stim- ulation. On the other hand, MS novel exams include cognitive assessment tests and three questionnaires, two of which are self-reported by patients. The “Cognitive Assessment" class includes exams to assess le- sions distribution, e.g. “Corpus Callosum Topography Score (CLTR)" and “Lesion Topography Score (LTS)", and tests for cognitive functions, e.g. “Symbol Digit Modalities Test (SDMT)" and “Paced Auditory Serial Addition Test (PASAT)". The questionnaires include the “Multiple Sclerosis Neuropsychological Question- naire (MSNQ)", “Fatigue Severity Score" (self-reported), and “Beck Depression Inventory-II (BDI-II)" (self- reported).
Figure 6: The Diagnostic Procedure area in HERO-Clinical. The components highlighted in orange and pink are novel diagnostic procedures or properties for MS and ALS patients respectively, included in HERO.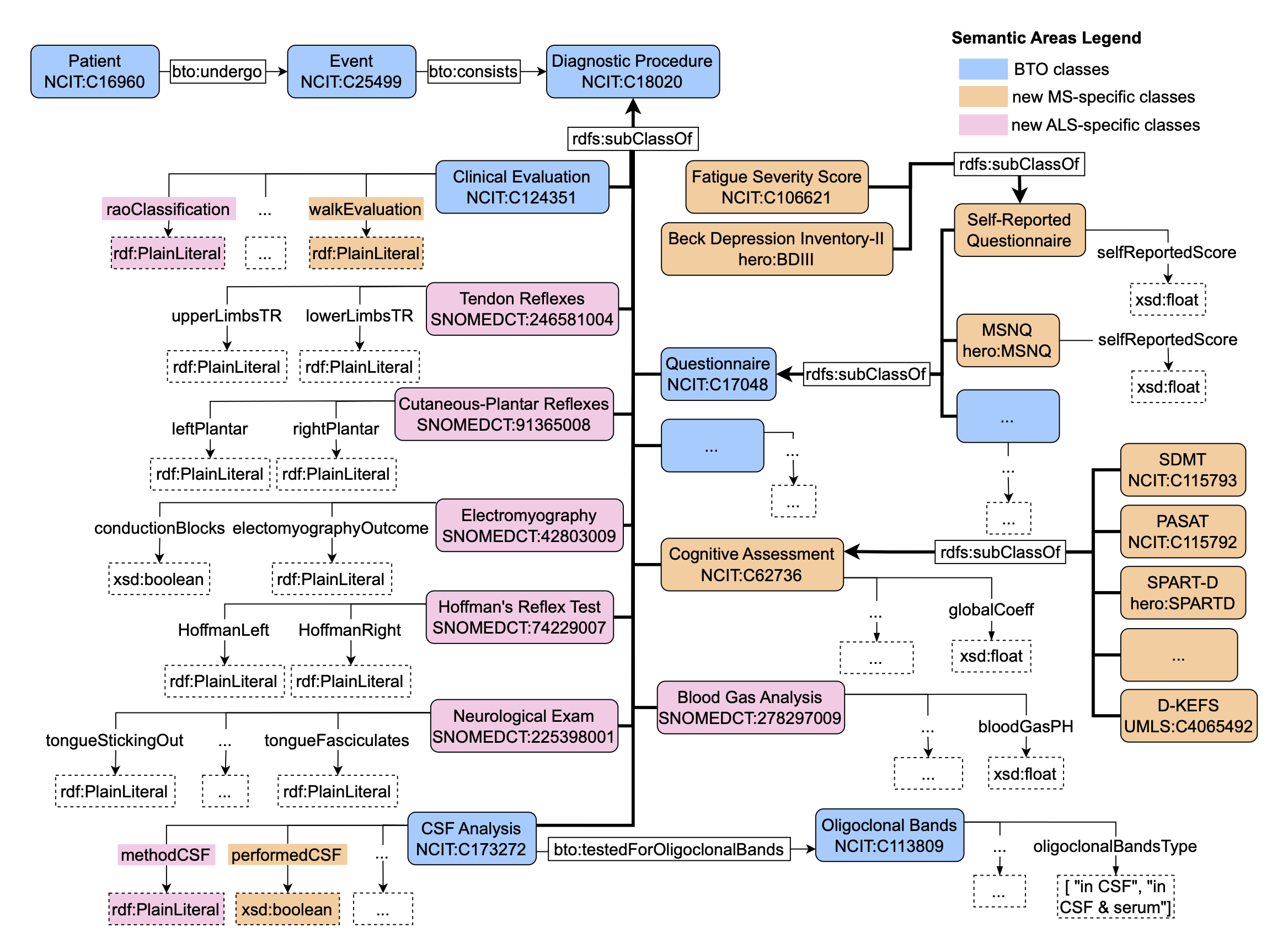
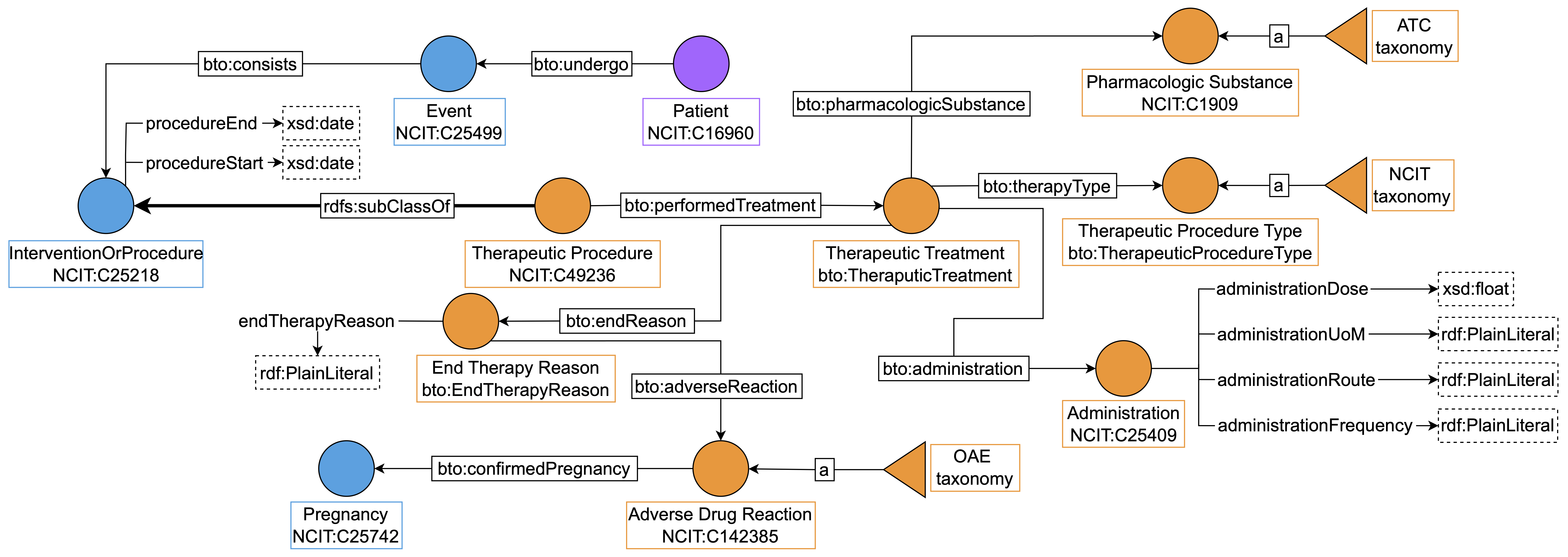
We identified a specific semantic field for “Therapeutic Procedures” which comprises
any treatment and the administration of any pharmacologic substance.
In a single therapeutic treatment, the clinician could administrate multiple phar-
macologic substances in different time periods. For this reason, we modeled this
area by introducing the class “Therapeutic Treatment”, which is the core of the
semantic area as illustrated by Figure 7 .
Figure 7: “Therapeutic Procedures” area. The core of the area is the “Therapeutic Treatment”
class, where one can store information about drug administrations and its specific aspects, such as
dose, frequency and end treatment. In particular, we modeled the “End Therapy Reason” as a
class so that we can link possible causes, such as adverse events and pregnancy.