The Hereditary Ontology for Genomics Data
- This version:
- https://w3id.org/hereditary/ontology/genomics/
- Latest version:
- https://w3id.org/hereditary/ontology/genomics/
- Ontology Version IRI:
- https://w3id.org/hereditary/ontology/genomics/1.0/
- Revision:
- 1.0
- Authors:
- Laura Menotti
- Gianmaria Silvello
- Download serialization:
-
- License:
- Cite as:
- Laura Menotti, Gianmaria Silvello. The Hereditary Ontology for Genomics Data. Revision: 1.0. Retrieved from: https://w3id.org/hereditary/ontology/genomics/schema/1.0/
Introduction back to ToC
The Genomic Data Modeling of the Hereditary Ontology (HERO) represents genomics data for ALS and MS patients. HERO-Clinical allows to store whether a patient’s genome presents specific gene mutations linked to ALS or MS, however, there is no representation for genetic testing and storing gene sequencing variations, e.g. Single Nucleotide Polymorphisms (SNPs).
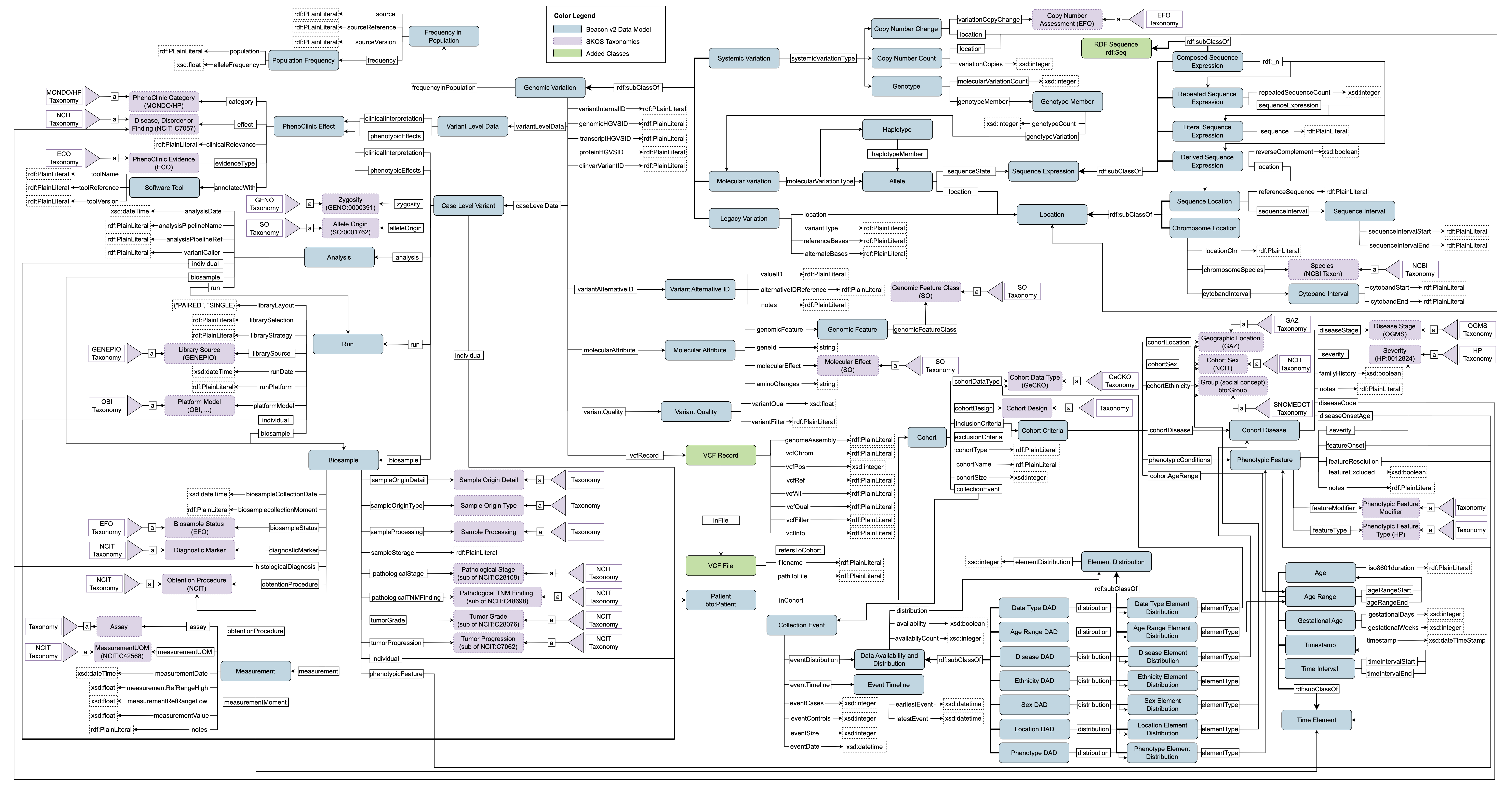
Figure 1 reports the complete schema of the Genomic Data Modeling. The Genomic Data Modeling in the HEREDITARY ontology follows the Beacon Data Model, employing the “Genomic Variation" component as the core class of the ontology. HERO-Genomics allows the storage of genetic variations, e.g. SNVs, insertions, deletions, and structural variants, alongside with their annotations, modeled with the “Case Level Variant" class. HERO-Genomics provides a direct mapping from VCFs files, or Beacon, to Resource Description Framework (RDF) format, thanks to classes “VCF Record", “VCF File", and “Variant Quality". It is possible to link HERO-Clinical to HERO-Genomics thanks to the shared “Patient" class. In addition, HERO-Genomics comprises the class “Disease, Disorder or Finding" defined in HERO-Clinical.
Namespace declarations
| [Ontology NS Prefix] | <https://w3id.org/hereditary/ontology/genomics/schema/> |
| bto | <https://w3id.org/brainteaser/ontology/schema/> |
| contact | <http://www.w3.org/2000/10/swap/pim/contact#> |
| dc | <http://purl.org/dc/elements/1.1/> |
| efo | <http://www.ebi.ac.uk/efo/> |
| foaf | <http://xmlns.com/foaf/0.1/> |
| obo | <http://purl.obolibrary.org/obo/> |
| orcid | <https://orcid.org/0000-0002-0676-682> |
| owl | <http://www.w3.org/2002/07/owl#> |
| rdf | <http://www.w3.org/1999/02/22-rdf-syntax-ns#> |
| rdfs | <http://www.w3.org/2000/01/rdf-schema#> |
| skos | <http://www.w3.org/2004/02/skos/core#> |
| terms | <http://purl.org/dc/terms/> |
| xml | <http://www.w3.org/XML/1998/namespace> |
| xsd | <http://www.w3.org/2001/XMLSchema#> |
Overview back to ToC
This ontology has the following classes and properties.Classes
- Age
- Age Range
- Age Range Data Availability and Distribution
- Age Range Element Distribution
- Allele
- Allele Origin
- Analysis
- Assay
- Biosample
- Biosample Status
- Case Level Variant
- Chromosome Location
- Cohort
- Cohort Criteria
- Cohort Data Type
- Cohort Design
- Cohort Disease
- Cohort Sex
- Collection Event
- Composed Sequence Expression
- Copy Number Assessment
- Copy Number Change
- Copy Number Count
- Cytoband Interval
- Data Availability and Distribution
- Data Type Data Availability and Distribution
- Data Type Element Distribution
- Derived Sequence Expression
- Diagnostic Marker
- Disease Data Availability and Distribution
- Disease Element Distribution
- Disease Stage
- Disease, Disorder or Finding
- Element Distribution
- Ethnicity Data Availability and Distribution
- Ethnicity Element Distribution
- Event Timeline
- Frequency In Population
- Genomic Feature
- Genomic Feature Class
- Genomic Variation
- Genotype
- Genotype Member
- Geographic Location
- Gestational Age
- Group (social concept)
- Haplotype
- Legacy Variation
- Library Source
- Literal Sequence Expression
- Location
- Location Data Availability and Distribution
- Location Element Distribution
- Measurement
- Molecular Attribute
- Molecular Effect
- Molecular Variation
- Obtention Procedure
- Pathological Stage
- Pathological TNM Finding
- Patient
- Person
- PhenoClinic Category
- PhenoClinic Effect
- PhenoClinic Evidence
- Phenotype Data Availability and Distribution
- Phenotype Element Distribution
- Phenotypic Feature
- Phenotypic Feature Modifier
- Phenotypic Feature Type
- Platform Model
- Population Frequency
- Repeated Sequence Expression
- Run
- Sample Origin Detail
- Sample Origin Type
- Sample Processing
- Sequence Expression
- Sequence Interval
- Sequence Location
- Severity
- Sex Data Availability and Distribution
- Sex Element Distribution
- Software Tool
- Species
- Systemic Variation
- Time Element
- Time Interval
- Timestamp
- Tumor Grade
- Tumor Progression
- Unit Of Measure
- Variant Alternative ID
- Variant Level Data
- Variant Quality
- VCF File
- VCF Record
- Zygosity
Object Properties
- ageRangeEnd
- ageRangeStart
- alleleOrigin
- analysis
- annotatedWith
- assay
- biosample
- biosampleStatus
- caseLevelData
- category
- chromosomeSpecies
- clinicalInterpretation
- cohortAgeRange
- cohortDataType
- cohortDesign
- cohortDisease
- cohortEthnicity
- cohortLocation
- cohortSex
- collectionEvent
- cytobandInterval
- diagnosticMarker
- diseaseAgeOnset
- diseaseCode
- diseaseStage
- distribution
- effect
- elementType
- eventDistribution
- eventTimeline
- evidence
- exclusionCriteria
- featureModifier
- featureOnset
- featureResolution
- featureType
- frequency
- frequencyInPopulation
- genomicFeature
- genomicFeatureClass
- genotypeMember
- genotypeVariation
- haplotypeMember
- histologicalDiagnosis
- inclusionCriteria
- inCohort
- individual
- inVcfFile
- librarySource
- location
- measurement
- measurementMoment
- measurementUOM
- molecularAttribute
- molecularEffect
- molecularVariationType
- obtentionProcedure
- pathologicalStage
- pathologicalTNMFinding
- phenotypicCondition
- phenotypicEffect
- phenotypicFeature
- platformModel
- refersToCohort
- run
- sampleOriginDetail
- sampleOriginType
- sampleProcessing
- sequenceComponent
- sequenceExpression
- sequenceInterval
- sequenceLocation
- sequenceState
- severity
- systemicVariationType
- timeIntervalEnd
- timeIntervalStart
- tumorGrade
- tumorProgression
- variantAlternativeID
- variantLevelData
- variantQuality
- variationCopyChange
- vcfRecord
- zygosity
Data Properties
- alleleFrequency
- alternateBases
- alternativeIDReference
- aminoacidChange
- analysisAligner
- analysisDate
- analysisPipelineName
- analysisPipelineRef
- availability
- availabilityCount
- biosampleCollectionDate
- biosampleCollectionMoment
- clinicalRelevance
- clinvarVariantID
- cohortName
- cohortSize
- cohortType
- cytobandEnd
- cytobandStart
- earliestEvent
- elementDistribution
- eventCases
- eventControls
- eventDate
- eventSize
- familyHistory
- featureExcluded
- filename
- filepath
- geneID
- genomeAssembly
- genomicHGVSID
- genotypeCount
- gestationalDays
- gestationalWeeks
- iso8601duration
- latestEvent
- libraryLayout
- librarySelection
- libraryStrategy
- locationChr
- measurementDate
- measurementRefRangeHigh
- measurementRefRangeLow
- measurementValue
- molecularVariationCount
- notes
- population
- proteinHGVSID
- referenceBases
- referenceSequence
- repeatedSequenceCount
- reverseComplement
- runDate
- runPlatform
- sampleStorage
- sequence
- sequenceIntervalEnd
- sequenceIntervalStart
- source
- sourceReference
- sourceVersion
- timestamp
- toolName
- toolReference
- toolVersion
- transcriptHGVSID
- valueID
- variantCaller
- variantFilter
- variantInternalID
- variantQual
- variantType
- variationCopies
- vcfAlt
- vcfChrom
- vcfFilter
- vcfInfo
- vcfPos
- vcfQual
- vcfRef
Named Individuals
- 5 Prime UTR Variant
- Abdominal Pain
- Abnormal Sample
- Acquired Hypothyroidism
- Acute Cystitis
- Acute Myocardial Infarction
- Acute Pyelonephritis
- Acute Respiratory Failure
- Acute Tonsillitis
- Adenocarcinoma
- Alanine Aminotransferase Increased
- Alcohol Dependance
- Allele Origin Taxonomy
- Allergic Asthma
- Allergic Rhinitis
- Alzheimer's Disease
- Amenorrhea
- Amenorrhea due to Pituitary Dysfunction
- Amyotrophic Lateral Sclerosis
- Anemia
- Angioedema
- Ankle Fracture
- Ann Arbor Stage IV Mantle Cell Lymphoma
- Ann Arbor Stage IV T-Cell Non-Hodgkin Lymphoma
- Anorectal Fistula
- Anorexia
- Anosmia
- Anxiety Disorder
- Arrhythmia
- Arthritis
- Asian (ethnic group)
- Aspiration Pneumonitis
- Assay Taxonomy
- Asthma
- Atopic Dermatitis
- Atrial Fibrillation
- Atrial Flutter
- Autoimmune Disease
- Autoimmune Nervous System Disorder
- Autoimmune Pancytopenia
- Bacterial Pneumonia
- Baker Cyst
- Basal Cell Carcinoma
- Behavioral Impairment
- Bell's Palsy
- Biosample Status Taxonomy
- Bipolar Disorder
- Black (ethnic group)
- Blepharitits
- Blepharospasm
- Borderline
- Bowel Obstruction
- Bradycardia
- Breast Carcinoma
- Brown-Sequard Syndrome
- Cancer Cell Line Sample
- Cancer TNM Finding Category
- Carcinoma
- Cardiovascular Disorder
- Cardiovascular System
- Case Control Design
- Caucasian
- Cavernous Hemangioma
- Celiac Disease
- Central Nervous System Degenerative Disorder
- Chicken Pox
- Chronic Lung Disorder
- Clinical Modifier
- Closed Dislocation of Shoulder
- Closed Fracture of Rib
- Closed Fracture of Tibia
- Cognitive Impariment
- Cohort Data Type Taxonomy
- Cohort Design Taxonomy
- Cohort Sex Taxonomy
- Cold Sore
- Colon Adenocarcinoma
- Common Bile Duct Stone
- Complete Genomic Deletion
- Compouns Heterozygous
- Congenital Adrenal Hyperplasia
- Copy Number Assessment
- Copy Number Assessment Taxonomy
- Copy Number Gain
- Copy Number Loss
- Coronary Artery Bypass Graft
- COVID-19 Infection
- Cranial Nerve VII Palsy
- Cutaneous Melanoma
- Cyst
- De-Novo Variant
- Deep Vein Thrombosis
- Dementia
- Depression
- Diabetes Mellitus
- Diabetic Ketoacidosis
- Diagnostic Marker Taxonomy
- Disease Grade Qualifier
- Disease or Disorder
- Disease or Disorder (MONDO)
- Disease Stage Qualifier
- Disease Stage Taxonomy
- Disease, Disorder or Finding Taxonomy
- DNA Sequencer
- Dog Bite
- Drug Hypersensitivity Syndrome
- Dyslipidemia
- Eczema
- Endometriosis
- Epidermal Inclusion Cyst
- Essential Hypertension
- Ethnic Group
- Experimental Evidence
- Extrapyramidal Disorder
- Favism
- Female
- Femur Fracture
- Fever Chills
- Fibroma
- Fibromyalgia
- Finding
- Flu-Like Symptoms
- Fungal Infection
- Gastritis
- Gastroenteritits
- Gastroesophageal Reflux
- Gastroplasty
- Generalized Epilepsy
- Genomic Feature Class Taxonomy
- Genomic Source
- Geographic Location Taxonomy
- Germline Variant
- Gianmaria Silvello
- Glaucoma
- Group Taxonomy
- Hashimoto Thyroiditis
- Head and Neck Finding
- Head Trauma
- Headache
- Heart Failure
- Hemizygous
- Hemizygous Insertion-Linked
- Hemizygous X-Linked
- Hemizygous Y-Linked
- Hepatic Toxicity
- Hepatitis
- Hepatitis A Infection
- Hepatitis B, Purified Antigen
- Hereditary Factor XI Deficiency
- Hereditary Hemolytic Anemia
- Hernia
- Herpes Simplex Keratitis
- Herpes Simplex Virus Infection
- Herpes Zoster
- Heterozygous
- High Grade Cervical Squamous Intraepithelial Neoplasia
- High-Level Copy Number Gain
- High-Level Copy Number Loss
- Hispanic
- Homozygous
- Human
- Hypersensitivity
- Hypertension
- Hypertensive Crisis
- Hyperthyroidism
- Hypothyroidism
- Infectious Mononucleosis
- Inferential Evidence
- Inguinal Hernia
- Injury
- Interstitial Pneumonia
- Intestinal Pseudo-Obstruction
- Iridocyclitis
- Iron-Deficiency Anemia
- Keratitis
- Keratoconus
- Klippel-Feil Syndrome
- Langerhans Cell Histiocytosis
- Laura Menotti
- Leukopenia
- Library Source
- Library Source Taxonomy
- Long QT Syndrome
- Low-Level Copy Number Gain
- Low-Level Copy Number Loss
- Lumbar Hernia
- Lumbar Puncture Headache
- Lymphopenia
- Male
- Malignant Colon Neoplasm
- Malignant Neoplasm
- Malignant Skin Neoplasm
- Malignant Thyroid Gland Neoplasm
- Material Entity
- Maternal Variant
- Melanocytic Nevus
- Metagenomic Source
- Metastasis Sample
- Metatranscriptomic Source
- Migraine
- Migraine With Aura
- Mild
- Missense Variant
- Moderate
- Molecular Effect Taxonomy
- Molluscum Contagiosum Virus
- Monoclonal B_Cell Lymphocytosis
- Monoclonal Gammopathy
- Mood Disorder
- Motor Neuron Disease
- Multiple Sclerosis
- Mumps
- Myasthenia Gravis
- Myeloproliferative Neoplasm
- Neck Trauma
- Neonatal Hearing Impairment
- Neoplasm
- Neoplasm by Special Category
- Neoplastic Sample
- Neurofibromatosis
- Neutropenic Disorder
- Non-Neoplastic Central Nervous System Disorder
- Nullizygous
- Obesity
- Obstructive Sleep Apnea Syndrome
- Obtention Procedure Taxonomy
- Opioid Use Disorder
- Optic Neuritis
- Osteoporosis
- Other Library Source
- Palmar Fibromatosis
- Pancytopenia
- Parkinson's Disease
- Paternal Variant
- Pathological Stage Taxonomy
- Pathological TNM Finding Taxonomy
- Pedigree Specific Variant
- Penicillin Allergy
- Personality Disorder
- Phenoclinic Category Taxonomy
- Phenoclinic Evidence Taxonomy
- Phenotypic Abnormality
- Phenotypic Feature Modifier Taxonomy
- Phenotypic Feature Type Taxonomy
- Pituitary Neuroendocrine Tumor
- Platform Model Taxonomy
- Pleural Effusion
- Pneumococcal Pneumonia
- Pneumonia
- Pneumonia Caused By SARS-CoV-2
- Population Specific Variant
- Post-Herpetic Neuralgia
- Primary Neoplasm
- Primary Progressive Aphasia
- Primary Tumor Sample
- Profound
- Psoriasis
- Psoriatic Arthritis
- Psychiatric Disorder
- Pulmonary Embolism
- Pulmonary Sarcoidosis
- Pulmonary Thromboembolism
- Radius Fracture
- Recurrent Tumor Sample
- Reference Sample
- Regional Base Ploidy
- Renal Colic
- Retinal Detachment
- Rheumatoid Arthritis
- Sample Dissociation
- Sample Origin Taxonomy
- Sample Origin Type Taxonomy
- Sample Processing Taxonomy
- Sarcoidosis
- Scarlet Fever
- Seizure Disorder
- Serositis
- Severe
- Severity
- Severity Taxonomy
- Shoulder Dislocation
- Sign
- Sign or Symptom
- Simple Heterozygous
- Sinusitis
- Skin Basal Cell Carcinoma
- Skin Basosquamous Cell Carcinoma
- Skin Carcinoma
- Skin Cyst
- Somatic Variant
- Species Taxonomy
- Spina Bifida
- Spinal and Bulbar Muscular Atrophy, X-linked 1
- Staphylococcal Infection
- Stop Gained NMD Escaping
- Stroke
- Study Design
- Subdural Hematoma
- Symptom
- Syncope
- Synthetic Source
- Thalassemia
- Thoracic Trauma
- Thromboembolism
- Thyroid Gland Disorder
- Thyroiditis
- Tibia Fracture
- Tinea Versicolor
- Tooth Abscess
- Toxoplasmosis
- Transcriptomic Source
- Transient Ischemic Attack
- Trigeminal Nerve Disorder
- Tumor Grade Taxonomy
- Tumor Progression Taxonomy
- Two Vessel Coronary Disease
- Type 1 Diabtes Mellitus
- Type 2 Diabetes Mellitus
- Unit by Category
- Unit of Measure Taxonomy
- Unknown
- Ureteral Stenosis
- Uterine Corpus Leiomyoma
- Variant Origin
- Vertigo
- Viral Pericarditis
- Viral RNA Source
- Vitiligo
- Vulvovaginal Candidiasis
- Vulvovaginitis
- Zygosity
- Zygosity Taxonomy
The Hereditary Ontology for Genomic Data: Description back to ToC
Domain requirements
In the context of the HEREDITARY project, genetic information is collected in VCF files comprising SNPs. A SNP is a one-letter place where an individual’s genome varies with respect to another sequence, usually called “Reference Genome". SNPs are SNVs present in a sufficiently large fraction, e.g. at least 1%, of a specific population.VCF is a text format widely used for storing genetic variations, such as Single Nucleotide Variants (SNVs), insertions, deletions, and structural variants, together with rich annotations [Danecek et al., 2011]. The format was developed for the 1000 Genome Project 7 and has been adopted in several other projects, e.g. dbSNP 8. Each VCF file comprises a header and a body. The former provides metadata describing the body of the file and keywords that optionally describe the fields used in the body. The latter is tap-separated and comprises 8 mandatory columns and an unlimited number of optional columns that may record additional information about the sample. When optional columns are used, the first of these columns describes the format of the additional columns. The mandatory columns are:
- CHROM, the name of the sequence, typically a chromosome, on which the variation is being called;
- POS, the one-based position of the variation;
- ID, the identifier of the variation, e.g. dbSNP rs identifier;
- REF, the reference base;
- ALT, the alternative allele;
- QUAL, the quality score associated with the inference of the given allele;
- FILTER, the flag indicating which filters have failed or PASS if all filters were successfully passed;
- INFO, the description of the variation. This column is highly variable and its content can vary across VCFs;
- FORMAT, an optional list of fields for describing the samples;
- SAMPLEs, optional values describing the samples.
The HEREDITARY ontology will answer the Beacon genomic queries provided and other queries of inter- est to the genomics community. The Beacon queries comprise:
- Sequence Queries for the existence of a specified sequence at a given genomic position;
- Range Queries for matching variants at least overlapping with a specified region;
- GeneId Queries for returning variants affecting a gene’s coding region;
- Bracket Queries for matching variants falling in a start range and end range;
- Genomic Allele Queries for matching variants with the specified allele;
- Amino Acid Change Queries for matching variants with the specified amino acid change.
Ontology Schema
Figure 2 reports the schema of genomic variations and VCF files. HERO- Genomics records systemic variations, e.g. Copy Number Change, molecular variations, and legacy varia- tions, e.g. SNPs. For each variation, one can store the VCF row with class “VCF Record" and link it to the corresponding file with class “VCF File". VCF annotations such as molecular effects and amino acid changes are represented by the class “Molecular Attribute". Class “Case Level Data" stores information about the ob- served zygosity, a SKOS taxonomy from the Gene Ontology, phenotypic effects, and clinical interpretations. In addition, the class links the genomic variation to the corresponding biosample (“Biosample"), run (“Run"), analysis (“Analysis"), and patient (“Patient" from HERO-Clinical). The location of the variation can stored by means of the class “Location", which provides a representation both for sequence location, to specify the sequence interval and reference genome, and chromosome location, to specify the cytoband interval of the chromosome where the variation occurs.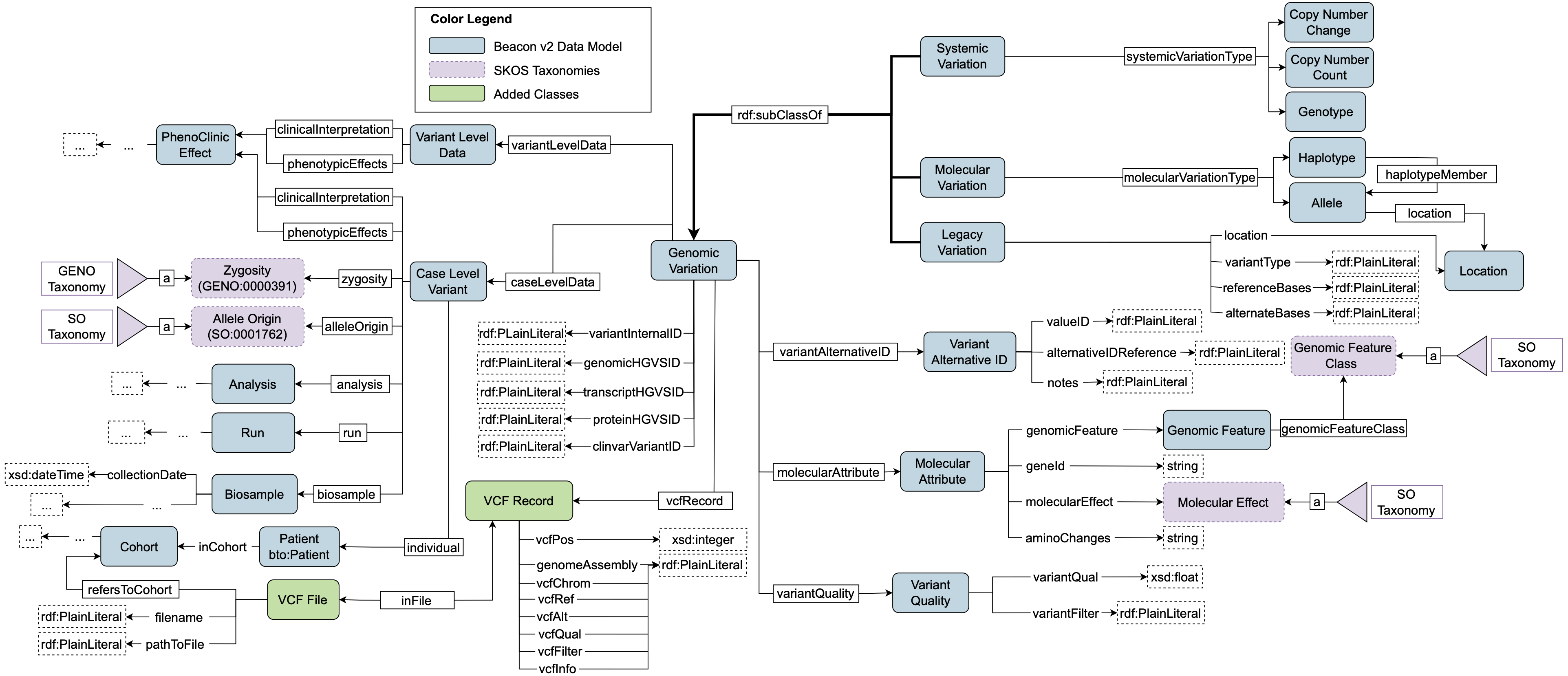
Cross-reference for classes, object properties and data properties back to ToC
This section provides details for each class and property defined by The Hereditary Ontology for Genomic Data.Classes
- Age
- Age Range
- Age Range Data Availability and Distribution
- Age Range Element Distribution
- Allele
- Allele Origin
- Analysis
- Assay
- Biosample
- Biosample Status
- Case Level Variant
- Chromosome Location
- Cohort
- Cohort Criteria
- Cohort Data Type
- Cohort Design
- Cohort Disease
- Cohort Sex
- Collection Event
- Composed Sequence Expression
- Copy Number Assessment
- Copy Number Change
- Copy Number Count
- Cytoband Interval
- Data Availability and Distribution
- Data Type Data Availability and Distribution
- Data Type Element Distribution
- Derived Sequence Expression
- Diagnostic Marker
- Disease Data Availability and Distribution
- Disease Element Distribution
- Disease Stage
- Disease, Disorder or Finding
- Element Distribution
- Ethnicity Data Availability and Distribution
- Ethnicity Element Distribution
- Event Timeline
- Frequency In Population
- Genomic Feature
- Genomic Feature Class
- Genomic Variation
- Genotype
- Genotype Member
- Geographic Location
- Gestational Age
- Group (social concept)
- Haplotype
- Legacy Variation
- Library Source
- Literal Sequence Expression
- Location
- Location Data Availability and Distribution
- Location Element Distribution
- Measurement
- Molecular Attribute
- Molecular Effect
- Molecular Variation
- Obtention Procedure
- Pathological Stage
- Pathological TNM Finding
- Patient
- Person
- PhenoClinic Category
- PhenoClinic Effect
- PhenoClinic Evidence
- Phenotype Data Availability and Distribution
- Phenotype Element Distribution
- Phenotypic Feature
- Phenotypic Feature Modifier
- Phenotypic Feature Type
- Platform Model
- Population Frequency
- Repeated Sequence Expression
- Run
- Sample Origin Detail
- Sample Origin Type
- Sample Processing
- Sequence Expression
- Sequence Interval
- Sequence Location
- Severity
- Sex Data Availability and Distribution
- Sex Element Distribution
- Software Tool
- Species
- Systemic Variation
- Time Element
- Time Interval
- Timestamp
- Tumor Grade
- Tumor Progression
- Unit Of Measure
- Variant Alternative ID
- Variant Level Data
- Variant Quality
- VCF File
- VCF Record
- Zygosity
Agec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/Age
- is equivalent to
- iso8601duration dp min 1
- has super-classes
- Time Element c
- is in domain of
- iso8601duration dp
- is in range of
- ageRangeEnd op, ageRangeStart op
Age Rangec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/AgeRange
- is equivalent to
- ageRangeEnd op min 1 Age c
- ageRangeStart op min 1 Age c
- has super-classes
- Time Element c
- is in domain of
- ageRangeEnd op, ageRangeStart op
- is in range of
- cohortAgeRange op
Age Range Data Availability and Distributionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/AgeRangeDAD
- is equivalent to
- distribution op some Age Range Element Distribution c
- has super-classes
- Data Availability and Distribution c
Age Range Element Distributionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/AgeRangeElementDistribution
- is equivalent to
- elementType op some Age Range c
- has super-classes
- Element Distribution c
Allelec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/Allele
- is in domain of
- sequenceState op
- is in range of
- haplotypeMember op
Allele Originc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/AlleleOrigin
- is in range of
- alleleOrigin op
- has members
- De-Novo Variant ni, Germline Variant ni, Maternal Variant ni, Paternal Variant ni, Pedigree Specific Variant ni, Population Specific Variant ni, Somatic Variant ni, Variant Origin ni
Analysisc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/Analysis
- is equivalent to
- analysisDate dp min 1
- analysisPipelineName dp min 1
- is in domain of
- analysisAligner dp, analysisDate dp, analysisPipelineName dp, analysisPipelineRef dp, variantCaller dp
- is in range of
- analysis op
Assayc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/Assay
- is in range of
- assay op
Biosamplec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/Biosample
- is equivalent to
- biosampleStatus op min 1 Biosample Status c
- sampleOriginType op min 1 Sample Origin Type c
- is in domain of
- biosampleCollectionDate dp, biosampleCollectionMoment dp, biosampleStatus op, diagnosticMarker op, histologicalDiagnosis op, measurement op, pathologicalStage op, pathologicalTNMFinding op, phenotypicFeature op, sampleOriginDetail op, sampleOriginType op, sampleProcessing op, sampleStorage dp, tumorGrade op, tumorProgression op
- is in range of
- biosample op
Biosample Statusc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/BiosampleStatus
- is in range of
- biosampleStatus op
- has members
- Abnormal Sample ni, Cancer Cell Line Sample ni, Case Control Design ni, Metastasis Sample ni, Neoplastic Sample ni, Primary Tumor Sample ni, Recurrent Tumor Sample ni, Reference Sample ni
Case Level Variantc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/CaseLevelVariant
- is equivalent to
- biosample op min 1 Biosample c
- is in domain of
- alleleOrigin op, analysis op, zygosity op
- is in range of
- caseLevelData op
Chromosome Locationc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/ChromosomeLocation
- is equivalent to
- chromosomeSpecies op exactly 1 Species c
- cytobandInterval op exactly 1 Cytoband Interval c
- locationChr dp exactly 1
- has super-classes
- Location c
- is in domain of
- chromosomeSpecies op, cytobandInterval op, locationChr dp
Cohortc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/Cohort
- is equivalent to
- cohortName dp exactly 1
- cohortType dp exactly 1
- is in domain of
- cohortDataType op, cohortDesign op, cohortName dp, cohortSize dp, cohortType dp, collectionEvent op, exclusionCriteria op, inclusionCriteria op
- is in range of
- inCohort op, refersToCohort op
Cohort Criteriac back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/CohortCriteria
- is in domain of
- cohortAgeRange op, cohortDisease op, cohortEthnicity op, cohortLocation op, cohortSex op, phenotypicCondition op
- is in range of
- exclusionCriteria op, inclusionCriteria op
Cohort Data Typec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/CohortDataType
- is in range of
- cohortDataType op
Cohort Designc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/CohortDesign
- is in range of
- cohortDesign op
- has members
- Study Design ni
Cohort Diseasec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/CohortDisease
- is in domain of
- diseaseAgeOnset op, diseaseCode op, diseaseStage op, familyHistory dp
- is in range of
- cohortDisease op
Cohort Sexc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/CohortSex
Collection Eventc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/CollectionEvent
- is in domain of
- eventCases dp, eventControls dp, eventDate dp, eventDistribution op, eventSize dp, eventTimeline op
- is in range of
- collectionEvent op
Composed Sequence Expressionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/ComposedSequenceExpression
- has super-classes
- Sequence Expression c, seq c
- is in domain of
- sequenceComponent op
Copy Number Assessmentc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/CopyNumberAssessment
- Is defined by
- http://www.ebi.ac.uk/efo/EFO_0030063
- is in range of
- variationCopyChange op
- has members
- Complete Genomic Deletion ni, Copy Number Assessment ni, Copy Number Gain ni, Copy Number Loss ni, High-Level Copy Number Gain ni, High-Level Copy Number Loss ni, Low-Level Copy Number Gain ni, Low-Level Copy Number Loss ni, Regional Base Ploidy ni
Copy Number Changec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/CopyNumberChange
- is equivalent to
- location op exactly 1 Location c
- variationCopyChange op exactly 1 Copy Number Assessment c
- is in domain of
- variationCopyChange op
Copy Number Countc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/CopyNumberCount
- is equivalent to
- location op exactly 1 Location c
- variationCopies dp exactly 1
- is in domain of
- variationCopies dp
Cytoband Intervalc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/CytobandInterval
- is equivalent to
- cytobandEnd dp exactly 1
- cytobandStart dp exactly 1
- is in domain of
- cytobandEnd dp, cytobandStart dp
- is in range of
- cytobandInterval op
Data Availability and Distributionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/DataAvailabilityAndDistribution
- is equivalent to
- availability dp exactly 1
- has sub-classes
- Age Range Data Availability and Distribution c, Data Type Data Availability and Distribution c, Disease Data Availability and Distribution c, Ethnicity Data Availability and Distribution c, Location Data Availability and Distribution c, Phenotype Data Availability and Distribution c, Sex Data Availability and Distribution c
- is in domain of
- availability dp, availabilityCount dp, distribution op
- is in range of
- eventDistribution op
Data Type Data Availability and Distributionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/DataTypeDAD
- is equivalent to
- distribution op some Data Type Element Distribution c
- has super-classes
- Data Availability and Distribution c
Data Type Element Distributionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/DataTypeElementDistribution
- is equivalent to
- elementType op some Cohort Data Type c
- has super-classes
- Element Distribution c
Derived Sequence Expressionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/DerivedSequenceExpression
- is equivalent to
- sequenceLocation op exactly 1 Sequence Location c
- reverseComplement dp exactly 1
- has super-classes
- Sequence Expression c
- is in domain of
- reverseComplement dp, sequenceLocation op
Diagnostic Markerc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/DiagnosticMarker
- is in range of
- diagnosticMarker op
Disease Data Availability and Distributionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/DiseaseDAD
- is equivalent to
- distribution op some Disease Element Distribution c
- has super-classes
- Data Availability and Distribution c
Disease Element Distributionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/DiseaseElementDistribution
- is equivalent to
- elementType op some Cohort Disease c
- has super-classes
- Element Distribution c
Disease Stagec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/DiseaseStage
- is in range of
- diseaseStage op
Element Distributionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/ElementDistribution
- has sub-classes
- Age Range Element Distribution c, Data Type Element Distribution c, Disease Element Distribution c, Ethnicity Element Distribution c, Location Element Distribution c, Phenotype Element Distribution c, Sex Element Distribution c
- is in domain of
- elementDistribution dp, elementType op
- is in range of
- distribution op
Ethnicity Data Availability and Distributionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/EthnicityDAD
- is equivalent to
- distribution op some Ethnicity Element Distribution c
- has super-classes
- Data Availability and Distribution c
Ethnicity Element Distributionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/EthnicityElementDistribution
- has super-classes
- Element Distribution c
Event Timelinec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/EventTimeline
- is in domain of
- earliestEvent dp, latestEvent dp
- is in range of
- eventTimeline op
Frequency In Populationc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/FrequencyInPopulation
- is equivalent to
- frequency op min 1 Population Frequency c
- source dp min 1
- sourceReference dp min 1
- is in domain of
- frequency op, source dp, sourceReference dp, sourceVersion dp
- is in range of
- frequencyInPopulation op
Genomic Featurec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/GenomicFeature
- is in domain of
- genomicFeatureClass op
- is in range of
- genomicFeature op
Genomic Feature Classc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/GenomicFeatureClass
- is equivalent to
- genomicFeatureClass op min 1 Genomic Feature Class c
- is in range of
- genomicFeatureClass op
- has members
- 5 Prime UTR Variant ni
Genomic Variationc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/GenomicVariation
- is equivalent to
- variantInternalID dp min 1
- has sub-classes
- Legacy Variation c, Molecular Variation c, Systemic Variation c
- is in domain of
- caseLevelData op, clinvarVariantID dp, frequencyInPopulation op, genomicHGVSID dp, molecularAttribute op, proteinHGVSID dp, transcriptHGVSID dp, variantAlternativeID op, variantInternalID dp, variantLevelData op, variantQuality op, vcfRecord op
Genotypec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/Genotype
- is equivalent to
- genotypeMember op min 1 Genotype Member c
- molecularVariationCount dp exactly 1
- is in domain of
- genotypeMember op, molecularVariationCount dp
Genotype Memberc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/GenotypeMember
- is equivalent to
- genotypeVariation op exactly 1 Molecular Variation c
- genotypeCount dp exactly 1
- is in domain of
- genotypeCount dp, genotypeVariation op
- is in range of
- genotypeMember op
Geographic Locationc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/GeographicLocation
- Is defined by
- http://purl.obolibrary.org/obo/GAZ_00000448
- is in range of
- cohortLocation op
Gestational Agec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/GestationalAge
- is equivalent to
- gestationalWeeks dp min 1
- has super-classes
- Time Element c
- is in domain of
- gestationalDays dp, gestationalWeeks dp
Haplotypec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/Haplotype
- is equivalent to
- haplotypeMember op min 2 Allele c
- is in domain of
- haplotypeMember op
Legacy Variationc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/LegacyVariation
- is equivalent to
- location op min 1 Location c
- alternateBases dp min 1
- variantType dp min 1
- has super-classes
- Genomic Variation c
- is in domain of
- alternateBases dp, referenceBases dp, variantType dp
Library Sourcec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/LibrarySource
- Is defined by
- http://purl.obolibrary.org/obo/GENEPIO_0001965
- is in range of
- librarySource op
- has members
- Genomic Source ni, Library Source ni, Metagenomic Source ni, Metatranscriptomic Source ni, Other Library Source ni, Synthetic Source ni, Transcriptomic Source ni, Viral RNA Source ni
Literal Sequence Expressionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/LiteralSequenceExpression
- is equivalent to
- sequence dp exactly 1
- has super-classes
- Sequence Expression c
- is in domain of
- sequence dp
Locationc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/Location
- has sub-classes
- Chromosome Location c, Sequence Location c
- is in range of
- location op
Location Data Availability and Distributionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/LocationDAD
- is equivalent to
- distribution op some Location Element Distribution c
- has super-classes
- Data Availability and Distribution c
Location Element Distributionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/LocationElementDistribution
- is equivalent to
- elementType op some Geographic Location c
- has super-classes
- Element Distribution c
Measurementc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/Measurement
- is equivalent to
- assay op min 1 Assay c
- measurementValue dp min 1
- is in domain of
- assay op, measurementDate dp, measurementMoment op, measurementRefRangeHigh dp, measurementRefRangeLow dp, measurementUOM op, measurementValue dp
- is in range of
- measurement op
Molecular Attributec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/MolecularAttribute
- is in domain of
- aminoacidChange dp, geneID dp, genomicFeature op, molecularEffect op
- is in range of
- molecularAttribute op
Molecular Effectc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/MolecularEffect
- is in range of
- molecularEffect op
- has members
- Missense Variant ni, Stop Gained NMD Escaping ni
Molecular Variationc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/MolecularVariation
- has super-classes
- Genomic Variation c
- is in domain of
- molecularVariationType op
- is in range of
- genotypeVariation op
Obtention Procedurec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/ObtentionProcedure
- Is defined by
- http://purl.obolibrary.org/obo/NCIT_C25218
- is in range of
- obtentionProcedure op
Pathological Stagec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/PathologicalStage
- is in range of
- pathologicalStage op
- has members
- Disease Stage Qualifier ni
Pathological TNM Findingc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/PathologicalTNMFinding
- Is defined by
- http://purl.obolibrary.org/obo/NCIT_C48698
- is in range of
- pathologicalTNMFinding op
- has members
- Cancer TNM Finding Category ni
Personc back to ToC or Class ToC
IRI: http://www.w3.org/2000/10/swap/pim/contact#Person
- has sub-classes
- Patient c
- is in range of
- individual op
- has members
- Gianmaria Silvello ni, Laura Menotti ni
PhenoClinic Categoryc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/PhenoClinicCategory
- is in range of
- category op
- has members
- Disease or Disorder (MONDO) ni, Phenotypic Abnormality ni
PhenoClinic Effectc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/PhenoClinicEffect
- is in domain of
- annotatedWith op, category op, clinicalRelevance dp, effect op, evidence op
- is in range of
- clinicalInterpretation op, phenotypicEffect op
PhenoClinic Evidencec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/PhenoClinicEvidence
- is in range of
- evidence op
- has members
- Experimental Evidence ni, Inferential Evidence ni
Phenotype Data Availability and Distributionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/PhenotypeDAD
- is equivalent to
- distribution op some Phenotype Element Distribution c
- has super-classes
- Data Availability and Distribution c
Phenotype Element Distributionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/PhenotypeElementDistribution
- is equivalent to
- elementType op some Phenotypic Feature c
- has super-classes
- Element Distribution c
Phenotypic Featurec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/PhenotypicFeature
- is equivalent to
- featureType op exactly 1 Phenotypic Feature Type c
- is in domain of
- featureExcluded dp, featureModifier op, featureOnset op, featureResolution op, featureType op
- is in range of
- phenotypicCondition op, phenotypicFeature op
Phenotypic Feature Modifierc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/PhenotypicFeatureModifier
- is in range of
- featureModifier op
- has members
- Clinical Modifier ni
Phenotypic Feature Typec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/PhenotypicFeatureType
- is in range of
- featureType op
Platform Modelc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/PlatformModel
- is in range of
- platformModel op
- has members
- DNA Sequencer ni
Population Frequencyc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/PopulationFrequency
- is equivalent to
- alleleFrequency dp min 1
- population dp min 1
- is in domain of
- alleleFrequency dp, population dp
- is in range of
- frequency op
Repeated Sequence Expressionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/RepeatedSequenceExpression
- has super-classes
- Sequence Expression c
- is in domain of
- repeatedSequenceCount dp, sequenceExpression op
Runc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/Run
- is equivalent to
- biosample op min 1 Biosample c
- runDate dp min 1
- is in domain of
- libraryLayout dp, librarySelection dp, librarySource op, libraryStrategy dp, platformModel op, runDate dp, runPlatform dp
- is in range of
- run op
Sample Origin Detailc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/SampleOriginDetail
- is in range of
- sampleOriginDetail op
Sample Origin Typec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/SampleOriginType
- is in range of
- sampleOriginType op
- has members
- Material Entity ni
Sample Processingc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/SampleProcessing
- is in range of
- sampleProcessing op
- has members
- Sample Dissociation ni
Sequence Expressionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/SequenceExpression
- has sub-classes
- Composed Sequence Expression c, Derived Sequence Expression c, Literal Sequence Expression c, Repeated Sequence Expression c
- is in range of
- sequenceState op
Sequence Intervalc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/SequenceInterval
- is equivalent to
- sequenceIntervalEnd dp exactly 1
- sequenceIntervalStart dp exactly 1
- is in domain of
- sequenceIntervalEnd dp, sequenceIntervalStart dp
- is in range of
- sequenceInterval op
Sequence Locationc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/SequenceLocation
- is equivalent to
- sequenceInterval op exactly 1 Sequence Interval c
- referenceSequence dp exactly 1
- has super-classes
- Location c
- is in domain of
- referenceSequence dp, sequenceInterval op
- is in range of
- sequenceLocation op
Severityc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/Severity
- Is defined by
- https://hpo.jax.org/browse/term/HP:0012824
Sex Data Availability and Distributionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/SexDAD
- is equivalent to
- distribution op some Sex Element Distribution c
- has super-classes
- Data Availability and Distribution c
Sex Element Distributionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/SexElementDistribution
- is equivalent to
- elementType op some Cohort Sex c
- has super-classes
- Element Distribution c
Software Toolc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/SoftwareTool
- is equivalent to
- toolName dp min 1
- toolReference dp min 1
- toolVersion dp min 1
- is in domain of
- toolName dp, toolReference dp, toolVersion dp
- is in range of
- annotatedWith op
Speciesc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/Species
- is in range of
- chromosomeSpecies op
- has members
- Human ni
Systemic Variationc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/SystemicVariation
- has super-classes
- Genomic Variation c
- is in domain of
- systemicVariationType op
Time Elementc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/TimeElement
- has sub-classes
- Age c, Age Range c, Gestational Age c, Time Interval c, Timestamp c
- is in range of
- diseaseAgeOnset op, featureOnset op, featureResolution op, measurementMoment op
Time Intervalc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/TimeInterval
- is equivalent to
- timeIntervalEnd op min 1 Timestamp c
- timeIntervalStart op min 1 Timestamp c
- has super-classes
- Time Element c
- is in domain of
- timeIntervalEnd op, timeIntervalStart op
Timestampc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/Timestamp
- is equivalent to
- timestamp dp min 1
- has super-classes
- Time Element c
- is in domain of
- timestamp dp
- is in range of
- timeIntervalEnd op, timeIntervalStart op
Tumor Gradec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/TumorGrade
- is in range of
- tumorGrade op
- has members
- Disease Grade Qualifier ni
Tumor Progressionc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/TumorProgression
- is in range of
- tumorProgression op
- has members
- Neoplasm by Special Category ni
Unit Of Measurec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/UnitOfMeasure
- Is defined by
- http://purl.obolibrary.org/obo/NCIT_C42568
- is in range of
- measurementUOM op
- has members
- Unit by Category ni
Variant Alternative IDc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/VariantAlternativeID
- is in domain of
- alternativeIDReference dp, valueID dp
- is in range of
- variantAlternativeID op
Variant Level Datac back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/VariantLevelData
- is in range of
- variantLevelData op
Variant Qualityc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/VariantQuality
- is in domain of
- variantFilter dp, variantQual dp
- is in range of
- variantQuality op
VCF Filec back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/VCFFile
- is in domain of
- filename dp, filepath dp, refersToCohort op
- is in range of
- inVcfFile op
VCF Recordc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/VCFRecord
Zygosityc back to ToC or Class ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/Zygosity
- is in range of
- zygosity op
- has members
- Compouns Heterozygous ni, Hemizygous ni, Hemizygous Insertion-Linked ni, Hemizygous X-Linked ni, Hemizygous Y-Linked ni, Heterozygous ni, Homozygous ni, Nullizygous ni, Simple Heterozygous ni, Zygosity ni
Object Properties
- ageRangeEnd
- ageRangeStart
- alleleOrigin
- analysis
- annotatedWith
- assay
- biosample
- biosampleStatus
- caseLevelData
- category
- chromosomeSpecies
- clinicalInterpretation
- cohortAgeRange
- cohortDataType
- cohortDesign
- cohortDisease
- cohortEthnicity
- cohortLocation
- cohortSex
- collectionEvent
- cytobandInterval
- diagnosticMarker
- diseaseAgeOnset
- diseaseCode
- diseaseStage
- distribution
- effect
- elementType
- eventDistribution
- eventTimeline
- evidence
- exclusionCriteria
- featureModifier
- featureOnset
- featureResolution
- featureType
- frequency
- frequencyInPopulation
- genomicFeature
- genomicFeatureClass
- genotypeMember
- genotypeVariation
- haplotypeMember
- histologicalDiagnosis
- inclusionCriteria
- inCohort
- individual
- inVcfFile
- librarySource
- location
- measurement
- measurementMoment
- measurementUOM
- molecularAttribute
- molecularEffect
- molecularVariationType
- obtentionProcedure
- pathologicalStage
- pathologicalTNMFinding
- phenotypicCondition
- phenotypicEffect
- phenotypicFeature
- platformModel
- refersToCohort
- run
- sampleOriginDetail
- sampleOriginType
- sampleProcessing
- sequenceComponent
- sequenceExpression
- sequenceInterval
- sequenceLocation
- sequenceState
- severity
- systemicVariationType
- timeIntervalEnd
- timeIntervalStart
- tumorGrade
- tumorProgression
- variantAlternativeID
- variantLevelData
- variantQuality
- variationCopyChange
- vcfRecord
- zygosity
ageRangeEndop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/ageRangeEnd
ageRangeStartop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/ageRangeStart
alleleOriginop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/alleleOrigin
- has domain
- Case Level Variant c
- has range
- Allele Origin c
analysisop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/analysis
- has domain
- Case Level Variant c
- has range
- Analysis c
annotatedWithop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/annotatedWith
- has domain
- PhenoClinic Effect c
- has range
- Software Tool c
assayop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/assay
- has domain
- Measurement c
- has range
- Assay c
biosampleop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/biosample
- has domain
- Analysis c or Case Level Variant c or Run c
- has range
- Biosample c
biosampleStatusop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/biosampleStatus
- has domain
- Biosample c
- has range
- Biosample Status c
caseLevelDataop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/caseLevelData
- has domain
- Genomic Variation c
- has range
- Case Level Variant c
categoryop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/category
- has domain
- PhenoClinic Effect c
- has range
- PhenoClinic Category c
chromosomeSpeciesop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/chromosomeSpecies
- has domain
- Chromosome Location c
- has range
- Species c
clinicalInterpretationop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/clinicalInterpretation
- has domain
- Case Level Variant c or Variant Level Data c
- has range
- PhenoClinic Effect c
cohortAgeRangeop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/cohortAgeRange
- has domain
- Cohort Criteria c
- has range
- Age Range c
cohortDataTypeop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/cohortDataType
- has domain
- Cohort c
- has range
- Cohort Data Type c
cohortDesignop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/cohortDesign
- has domain
- Cohort c
- has range
- Cohort Design c
cohortDiseaseop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/cohortDisease
- has domain
- Cohort Criteria c
- has range
- Cohort Disease c
cohortEthnicityop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/cohortEthnicity
- has domain
- Cohort Criteria c
cohortLocationop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/cohortLocation
- has domain
- Cohort Criteria c
- has range
- Geographic Location c
cohortSexop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/cohortSex
- has domain
- Cohort Criteria c
- has range
- Cohort Sex c
collectionEventop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/collectionEvent
- has domain
- Cohort c
- has range
- Collection Event c
cytobandIntervalop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/cytobandInterval
- has domain
- Chromosome Location c
- has range
- Cytoband Interval c
diagnosticMarkerop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/diagnosticMarker
- has domain
- Biosample c
- has range
- Diagnostic Marker c
diseaseAgeOnsetop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/diseaseAgeOnset
- has domain
- Cohort Disease c
- has range
- Time Element c
diseaseCodeop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/diseaseCode
- has domain
- Cohort Disease c
diseaseStageop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/diseaseStage
- has domain
- Cohort Disease c
- has range
- Disease Stage c
distributionop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/distribution
- has domain
- Data Availability and Distribution c
- has range
- Element Distribution c
effectop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/effect
- has domain
- PhenoClinic Effect c
elementTypeop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/elementType
- has domain
- Element Distribution c
eventDistributionop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/eventDistribution
- has domain
- Collection Event c
- has range
- Data Availability and Distribution c
eventTimelineop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/eventTimeline
- has domain
- Collection Event c
- has range
- Event Timeline c
evidenceop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/evidence
- has domain
- PhenoClinic Effect c
- has range
- PhenoClinic Evidence c
exclusionCriteriaop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/exclusionCriteria
- has domain
- Cohort c
- has range
- Cohort Criteria c
featureModifierop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/featureModifier
- has domain
- Phenotypic Feature c
- has range
- Phenotypic Feature Modifier c
featureOnsetop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/featureOnset
- has domain
- Phenotypic Feature c
- has range
- Time Element c
featureResolutionop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/featureResolution
- has domain
- Phenotypic Feature c
- has range
- Time Element c
featureTypeop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/featureType
- has domain
- Phenotypic Feature c
- has range
- Phenotypic Feature Type c
frequencyop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/frequency
- has domain
- Frequency In Population c
- has range
- Population Frequency c
frequencyInPopulationop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/frequencyInPopulation
- has domain
- Genomic Variation c
- has range
- Frequency In Population c
genomicFeatureop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/genomicFeature
- has domain
- Molecular Attribute c
- has range
- Genomic Feature c
genomicFeatureClassop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/genomicFeatureClass
- has domain
- Genomic Feature c
- has range
- Genomic Feature Class c
genotypeMemberop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/genotypeMember
- has domain
- Genotype c
- has range
- Genotype Member c
genotypeVariationop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/genotypeVariation
- has domain
- Genotype Member c
- has range
- Molecular Variation c
haplotypeMemberop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/haplotypeMember
histologicalDiagnosisop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/histologicalDiagnosis
- has domain
- Biosample c
inclusionCriteriaop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/inclusionCriteria
- has domain
- Cohort c
- has range
- Cohort Criteria c
inCohortop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/inCohort
individualop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/individual
- has domain
- Analysis c or Biosample c or Case Level Variant c or Run c
- has range
- Person c
inVcfFileop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/inVcfFile
- has domain
- VCF Record c
- has range
- VCF File c
librarySourceop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/librarySource
- has domain
- Run c
- has range
- Library Source c
locationop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/location
- has domain
- Allele c or Copy Number Change c or Copy Number Count c or Legacy Variation c
- has range
- Location c
measurementop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/measurement
- has domain
- Biosample c
- has range
- Measurement c
measurementMomentop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/measurementMoment
- has domain
- Measurement c
- has range
- Time Element c
measurementUOMop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/measurementUOM
- has domain
- Measurement c
- has range
- Unit Of Measure c
molecularAttributeop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/molecularAttributes
- has domain
- Genomic Variation c
- has range
- Molecular Attribute c
molecularEffectop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/molecularEffect
- has domain
- Molecular Attribute c
- has range
- Molecular Effect c
molecularVariationTypeop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/molecularVariationType
has characteristics: functional
- has domain
- Molecular Variation c
- has range
- Allele c or Haplotype c
obtentionProcedureop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/obtentionProcedure
- has domain
- Biosample c or Measurement c
- has range
- Obtention Procedure c
pathologicalStageop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/pathologicalStage
- has domain
- Biosample c
- has range
- Pathological Stage c
pathologicalTNMFindingop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/pathologicalTNMFinding
- has domain
- Biosample c
- has range
- Pathological TNM Finding c
phenotypicConditionop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/phenotypicConditions
- has domain
- Cohort Criteria c
- has range
- Phenotypic Feature c
phenotypicEffectop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/phenotypicEffect
- has domain
- Case Level Variant c or Variant Level Data c
- has range
- PhenoClinic Effect c
phenotypicFeatureop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/phenotypicFeature
- has domain
- Biosample c
- has range
- Phenotypic Feature c
platformModelop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/platformModel
- has domain
- Run c
- has range
- Platform Model c
refersToCohortop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/refersToCohort
runop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/run
- has domain
- Analysis c or Case Level Variant c
- has range
- Run c
sampleOriginDetailop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/sampleOriginDetail
- has domain
- Biosample c
- has range
- Sample Origin Detail c
sampleOriginTypeop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/sampleOriginType
- has domain
- Biosample c
- has range
- Sample Origin Type c
sampleProcessingop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/sampleProcessing
- has domain
- Biosample c
- has range
- Sample Processing c
sequenceComponentop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/sequenceComponent
- has domain
- Composed Sequence Expression c
- has range
- Derived Sequence Expression c or Literal Sequence Expression c or Repeated Sequence Expression c
sequenceExpressionop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/sequenceExpression
- has domain
- Repeated Sequence Expression c
- has range
- Derived Sequence Expression c or Literal Sequence Expression c
sequenceIntervalop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/sequenceInterval
- has domain
- Sequence Location c
- has range
- Sequence Interval c
sequenceLocationop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/sequenceLocation
- has domain
- Derived Sequence Expression c
- has range
- Sequence Location c
sequenceStateop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/sequenceState
- has domain
- Allele c
- has range
- Sequence Expression c
severityop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/severity
- has domain
- Cohort Disease c or Phenotypic Feature c
- has range
- Severity c
systemicVariationTypeop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/systemicVariationType
has characteristics: functional
- has domain
- Systemic Variation c
- has range
- Copy Number Change c or Copy Number Count c or Genotype c
timeIntervalEndop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/timeIntervalEnd
- has domain
- Time Interval c
- has range
- Timestamp c
timeIntervalStartop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/timeIntervalStart
- has domain
- Time Interval c
- has range
- Timestamp c
tumorGradeop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/tumorGrade
- has domain
- Biosample c
- has range
- Tumor Grade c
tumorProgressionop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/tumorProgression
- has domain
- Biosample c
- has range
- Tumor Progression c
variantAlternativeIDop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/variantAlternativeID
- has domain
- Genomic Variation c
- has range
- Variant Alternative ID c
variantLevelDataop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/variantLevelData
- has domain
- Genomic Variation c
- has range
- Variant Level Data c
variantQualityop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/variantQuality
- has domain
- Genomic Variation c
- has range
- Variant Quality c
variationCopyChangeop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/variationCopyChange
- has domain
- Copy Number Change c
- has range
- Copy Number Assessment c
vcfRecordop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/vcfRecord
- has domain
- Genomic Variation c
- has range
- VCF Record c
zygosityop back to ToC or Object Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/zygosity
- has domain
- Case Level Variant c
- has range
- Zygosity c
Data Properties
- alleleFrequency
- alternateBases
- alternativeIDReference
- aminoacidChange
- analysisAligner
- analysisDate
- analysisPipelineName
- analysisPipelineRef
- availability
- availabilityCount
- biosampleCollectionDate
- biosampleCollectionMoment
- clinicalRelevance
- clinvarVariantID
- cohortName
- cohortSize
- cohortType
- cytobandEnd
- cytobandStart
- earliestEvent
- elementDistribution
- eventCases
- eventControls
- eventDate
- eventSize
- familyHistory
- featureExcluded
- filename
- filepath
- geneID
- genomeAssembly
- genomicHGVSID
- genotypeCount
- gestationalDays
- gestationalWeeks
- iso8601duration
- latestEvent
- libraryLayout
- librarySelection
- libraryStrategy
- locationChr
- measurementDate
- measurementRefRangeHigh
- measurementRefRangeLow
- measurementValue
- molecularVariationCount
- notes
- population
- proteinHGVSID
- referenceBases
- referenceSequence
- repeatedSequenceCount
- reverseComplement
- runDate
- runPlatform
- sampleStorage
- sequence
- sequenceIntervalEnd
- sequenceIntervalStart
- source
- sourceReference
- sourceVersion
- timestamp
- toolName
- toolReference
- toolVersion
- transcriptHGVSID
- valueID
- variantCaller
- variantFilter
- variantInternalID
- variantQual
- variantType
- variationCopies
- vcfAlt
- vcfChrom
- vcfFilter
- vcfInfo
- vcfPos
- vcfQual
- vcfRef
alleleFrequencydp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/alleleFrequency
- has domain
- Population Frequency c
- has range
- float
alternateBasesdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/alternateBases
- has domain
- Legacy Variation c
- has range
- plain literal
alternativeIDReferencedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/alternativeIDReference
- has domain
- Variant Alternative ID c
- has range
- plain literal
aminoacidChangedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/aminoacidChange
- has domain
- Molecular Attribute c
- has range
- plain literal
analysisAlignerdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/analysisAligner
- has domain
- Analysis c
- has range
- plain literal
analysisDatedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/analysisDate
analysisPipelineNamedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/analysisPipelineName
- has domain
- Analysis c
- has range
- plain literal
analysisPipelineRefdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/analysisPipelineRef
- has domain
- Analysis c
- has range
- plain literal
availabilitydp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/availability
- has domain
- Data Availability and Distribution c
- has range
- boolean
availabilityCountdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/availabilityCount
- has domain
- Data Availability and Distribution c
- has range
- integer
biosampleCollectionDatedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/biosampleCollectionDate
biosampleCollectionMomentdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/biosampleCollectionMoment
- has domain
- Biosample c
- has range
- plain literal
clinicalRelevancedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/clinicalRelevance
- has domain
- PhenoClinic Effect c
- has range
- { "Benign" , "Likely Benign" , "Likely Pathogenic" , "Pathogenic" , "Uncertain Significance" }
clinvarVariantIDdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/clinvarVariantID
- has domain
- Genomic Variation c
- has range
- plain literal
cohortNamedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/cohortName
- has domain
- Cohort c
- has range
- plain literal
cohortSizedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/cohortSize
cohortTypedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/cohortType
- has domain
- Cohort c
- has range
- plain literal
cytobandEnddp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/cytobandEnd
- has domain
- Cytoband Interval c
- has range
- plain literal
cytobandStartdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/cytobandStart
- has domain
- Cytoband Interval c
- has range
- plain literal
earliestEventdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/earliestEvent
- has domain
- Event Timeline c
- has range
- date time
elementDistributiondp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/elementDistribution
- has domain
- Element Distribution c
- has range
- integer
eventCasesdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/eventCases
- has domain
- Collection Event c
- has range
- integer
eventControlsdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/eventControls
- has domain
- Collection Event c
- has range
- integer
eventDatedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/eventDate
- has domain
- Collection Event c
- has range
- date time
eventSizedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/eventSize
- has domain
- Collection Event c
- has range
- integer
familyHistorydp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/familyHistory
- has domain
- Cohort Disease c
- has range
- boolean
featureExcludeddp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/featureExcluded
- has domain
- Phenotypic Feature c
- has range
- boolean
filenamedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/filename
- has domain
- VCF File c
- has range
- plain literal
filepathdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/filepath
- has domain
- VCF File c
- has range
- plain literal
geneIDdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/geneID
- has domain
- Molecular Attribute c
- has range
- plain literal
genomeAssemblydp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/genomeAssembly
- has domain
- VCF Record c
- has range
- plain literal
genomicHGVSIDdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/genomicHGVSID
- has domain
- Genomic Variation c
- has range
- plain literal
genotypeCountdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/genotypeCount
- has domain
- Genotype Member c
- has range
- integer
gestationalDaysdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/gestationalDays
- has domain
- Gestational Age c
- has range
- integer
gestationalWeeksdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/gestationalWeeks
- has domain
- Gestational Age c
- has range
- integer
iso8601durationdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/iso8601duration
- has domain
- Age c
- has range
- plain literal
latestEventdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/latestEvent
- has domain
- Event Timeline c
- has range
- date time
libraryLayoutdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/libraryLayout
- has domain
- Run c
- has range
- { "PAIRED" , "SINGLE" }
librarySelectiondp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/librarySelection
- has domain
- Run c
- has range
- plain literal
libraryStrategydp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/libraryStrategy
- has domain
- Run c
- has range
- plain literal
locationChrdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/locationChr
- has domain
- Chromosome Location c
- has range
- plain literal
measurementDatedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/measurementDate
- has domain
- Measurement c
- has range
- date time
measurementRefRangeHighdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/measurementRefRangeHigh
- has domain
- Measurement c
- has range
- float
measurementRefRangeLowdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/measurementRefRangeLow
- has domain
- Measurement c
- has range
- float
measurementValuedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/measurementValue
- has domain
- Measurement c
- has range
- float
molecularVariationCountdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/molecularVariationCount
notesdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/notes
- has domain
- Cohort Disease c or Measurement c or Phenotypic Feature c or Variant Alternative ID c
- has range
- plain literal
populationdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/population
- has domain
- Population Frequency c
- has range
- plain literal
proteinHGVSIDdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/proteinHGVSID
- has domain
- Genomic Variation c
- has range
- plain literal
referenceBasesdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/referenceBases
- has domain
- Legacy Variation c
- has range
- plain literal
referenceSequencedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/referenceSequence
- has domain
- Sequence Location c
- has range
- plain literal
repeatedSequenceCountdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/repeatedSequenceCount
- has domain
- Repeated Sequence Expression c
- has range
- integer
reverseComplementdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/reverseComplement
- has domain
- Derived Sequence Expression c
- has range
- boolean
runDatedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/runDate
runPlatformdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/runPlatform
- has domain
- Run c
- has range
- plain literal
sampleStoragedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/sampleStorage
- has domain
- Biosample c
- has range
- plain literal
sequencedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/sequence
- has domain
- Literal Sequence Expression c
- has range
- plain literal
sequenceIntervalEnddp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/sequenceIntervalEnd
- has domain
- Sequence Interval c
- has range
- plain literal
sequenceIntervalStartdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/sequenceIntervalStart
- has domain
- Sequence Interval c
- has range
- plain literal
sourcedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/source
- has domain
- Frequency In Population c
- has range
- plain literal
sourceReferencedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/sourceReference
- has domain
- Frequency In Population c
- has range
- plain literal
sourceVersiondp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/sourceVersion
- has domain
- Frequency In Population c
- has range
- plain literal
timestampdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/timestamp
- has domain
- Timestamp c
- has range
- date time stamp
toolNamedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/toolName
- has domain
- Software Tool c
- has range
- plain literal
toolReferencedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/toolReference
- has domain
- Software Tool c
- has range
- plain literal
toolVersiondp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/toolVersion
- has domain
- Software Tool c
- has range
- plain literal
transcriptHGVSIDdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/transcriptHGVSID
- has domain
- Genomic Variation c
- has range
- plain literal
valueIDdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/valueID
- has domain
- Variant Alternative ID c
- has range
- plain literal
variantCallerdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/variantCaller
- has domain
- Analysis c
- has range
- plain literal
variantFilterdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/variantFilter
- has domain
- Variant Quality c
- has range
- plain literal
variantInternalIDdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/variantInternalID
- has domain
- Genomic Variation c
- has range
- plain literal
variantQualdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/variantQual
- has domain
- Variant Quality c
- has range
- float
variantTypedp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/variantType
- has domain
- Legacy Variation c
- has range
- plain literal
variationCopiesdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/variationCopies
- has domain
- Copy Number Count c
- has range
- integer
vcfAltdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/vcfAlt
- has domain
- VCF Record c
- has range
- plain literal
vcfChromdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/vcfChrom
- has domain
- VCF Record c
- has range
- plain literal
vcfFilterdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/vcfFilter
- has domain
- VCF Record c
- has range
- plain literal
vcfInfodp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/vcfInfo
- has domain
- VCF Record c
- has range
- plain literal
vcfPosdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/vcfPos
- has domain
- VCF Record c
- has range
- plain literal
vcfQualdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/vcfQual
- has domain
- VCF Record c
- has range
- plain literal
vcfRefdp back to ToC or Data Property ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/vcfRef
- has domain
- VCF Record c
- has range
- plain literal
Named Individuals
- 5 Prime UTR Variant
- Abdominal Pain
- Abnormal Sample
- Acquired Hypothyroidism
- Acute Cystitis
- Acute Myocardial Infarction
- Acute Pyelonephritis
- Acute Respiratory Failure
- Acute Tonsillitis
- Adenocarcinoma
- Alanine Aminotransferase Increased
- Alcohol Dependance
- Allele Origin Taxonomy
- Allergic Asthma
- Allergic Rhinitis
- Alzheimer's Disease
- Amenorrhea
- Amenorrhea due to Pituitary Dysfunction
- Amyotrophic Lateral Sclerosis
- Anemia
- Angioedema
- Ankle Fracture
- Ann Arbor Stage IV Mantle Cell Lymphoma
- Ann Arbor Stage IV T-Cell Non-Hodgkin Lymphoma
- Anorectal Fistula
- Anorexia
- Anosmia
- Anxiety Disorder
- Arrhythmia
- Arthritis
- Asian (ethnic group)
- Aspiration Pneumonitis
- Assay Taxonomy
- Asthma
- Atopic Dermatitis
- Atrial Fibrillation
- Atrial Flutter
- Autoimmune Disease
- Autoimmune Nervous System Disorder
- Autoimmune Pancytopenia
- Bacterial Pneumonia
- Baker Cyst
- Basal Cell Carcinoma
- Behavioral Impairment
- Bell's Palsy
- Biosample Status Taxonomy
- Bipolar Disorder
- Black (ethnic group)
- Blepharitits
- Blepharospasm
- Borderline
- Bowel Obstruction
- Bradycardia
- Breast Carcinoma
- Brown-Sequard Syndrome
- Cancer Cell Line Sample
- Cancer TNM Finding Category
- Carcinoma
- Cardiovascular Disorder
- Cardiovascular System
- Case Control Design
- Caucasian
- Cavernous Hemangioma
- Celiac Disease
- Central Nervous System Degenerative Disorder
- Chicken Pox
- Chronic Lung Disorder
- Clinical Modifier
- Closed Dislocation of Shoulder
- Closed Fracture of Rib
- Closed Fracture of Tibia
- Cognitive Impariment
- Cohort Data Type Taxonomy
- Cohort Design Taxonomy
- Cohort Sex Taxonomy
- Cold Sore
- Colon Adenocarcinoma
- Common Bile Duct Stone
- Complete Genomic Deletion
- Compouns Heterozygous
- Congenital Adrenal Hyperplasia
- Copy Number Assessment
- Copy Number Assessment Taxonomy
- Copy Number Gain
- Copy Number Loss
- Coronary Artery Bypass Graft
- COVID-19 Infection
- Cranial Nerve VII Palsy
- Cutaneous Melanoma
- Cyst
- De-Novo Variant
- Deep Vein Thrombosis
- Dementia
- Depression
- Diabetes Mellitus
- Diabetic Ketoacidosis
- Diagnostic Marker Taxonomy
- Disease Grade Qualifier
- Disease or Disorder
- Disease or Disorder (MONDO)
- Disease Stage Qualifier
- Disease Stage Taxonomy
- Disease, Disorder or Finding Taxonomy
- DNA Sequencer
- Dog Bite
- Drug Hypersensitivity Syndrome
- Dyslipidemia
- Eczema
- Endometriosis
- Epidermal Inclusion Cyst
- Essential Hypertension
- Ethnic Group
- Experimental Evidence
- Extrapyramidal Disorder
- Favism
- Female
- Femur Fracture
- Fever Chills
- Fibroma
- Fibromyalgia
- Finding
- Flu-Like Symptoms
- Fungal Infection
- Gastritis
- Gastroenteritits
- Gastroesophageal Reflux
- Gastroplasty
- Generalized Epilepsy
- Genomic Feature Class Taxonomy
- Genomic Source
- Geographic Location Taxonomy
- Germline Variant
- Gianmaria Silvello
- Glaucoma
- Group Taxonomy
- Hashimoto Thyroiditis
- Head and Neck Finding
- Head Trauma
- Headache
- Heart Failure
- Hemizygous
- Hemizygous Insertion-Linked
- Hemizygous X-Linked
- Hemizygous Y-Linked
- Hepatic Toxicity
- Hepatitis
- Hepatitis A Infection
- Hepatitis B, Purified Antigen
- Hereditary Factor XI Deficiency
- Hereditary Hemolytic Anemia
- Hernia
- Herpes Simplex Keratitis
- Herpes Simplex Virus Infection
- Herpes Zoster
- Heterozygous
- High Grade Cervical Squamous Intraepithelial Neoplasia
- High-Level Copy Number Gain
- High-Level Copy Number Loss
- Hispanic
- Homozygous
- Human
- Hypersensitivity
- Hypertension
- Hypertensive Crisis
- Hyperthyroidism
- Hypothyroidism
- Infectious Mononucleosis
- Inferential Evidence
- Inguinal Hernia
- Injury
- Interstitial Pneumonia
- Intestinal Pseudo-Obstruction
- Iridocyclitis
- Iron-Deficiency Anemia
- Keratitis
- Keratoconus
- Klippel-Feil Syndrome
- Langerhans Cell Histiocytosis
- Laura Menotti
- Leukopenia
- Library Source
- Library Source Taxonomy
- Long QT Syndrome
- Low-Level Copy Number Gain
- Low-Level Copy Number Loss
- Lumbar Hernia
- Lumbar Puncture Headache
- Lymphopenia
- Male
- Malignant Colon Neoplasm
- Malignant Neoplasm
- Malignant Skin Neoplasm
- Malignant Thyroid Gland Neoplasm
- Material Entity
- Maternal Variant
- Melanocytic Nevus
- Metagenomic Source
- Metastasis Sample
- Metatranscriptomic Source
- Migraine
- Migraine With Aura
- Mild
- Missense Variant
- Moderate
- Molecular Effect Taxonomy
- Molluscum Contagiosum Virus
- Monoclonal B_Cell Lymphocytosis
- Monoclonal Gammopathy
- Mood Disorder
- Motor Neuron Disease
- Multiple Sclerosis
- Mumps
- Myasthenia Gravis
- Myeloproliferative Neoplasm
- Neck Trauma
- Neonatal Hearing Impairment
- Neoplasm
- Neoplasm by Special Category
- Neoplastic Sample
- Neurofibromatosis
- Neutropenic Disorder
- Non-Neoplastic Central Nervous System Disorder
- Nullizygous
- Obesity
- Obstructive Sleep Apnea Syndrome
- Obtention Procedure Taxonomy
- Opioid Use Disorder
- Optic Neuritis
- Osteoporosis
- Other Library Source
- Palmar Fibromatosis
- Pancytopenia
- Parkinson's Disease
- Paternal Variant
- Pathological Stage Taxonomy
- Pathological TNM Finding Taxonomy
- Pedigree Specific Variant
- Penicillin Allergy
- Personality Disorder
- Phenoclinic Category Taxonomy
- Phenoclinic Evidence Taxonomy
- Phenotypic Abnormality
- Phenotypic Feature Modifier Taxonomy
- Phenotypic Feature Type Taxonomy
- Pituitary Neuroendocrine Tumor
- Platform Model Taxonomy
- Pleural Effusion
- Pneumococcal Pneumonia
- Pneumonia
- Pneumonia Caused By SARS-CoV-2
- Population Specific Variant
- Post-Herpetic Neuralgia
- Primary Neoplasm
- Primary Progressive Aphasia
- Primary Tumor Sample
- Profound
- Psoriasis
- Psoriatic Arthritis
- Psychiatric Disorder
- Pulmonary Embolism
- Pulmonary Sarcoidosis
- Pulmonary Thromboembolism
- Radius Fracture
- Recurrent Tumor Sample
- Reference Sample
- Regional Base Ploidy
- Renal Colic
- Retinal Detachment
- Rheumatoid Arthritis
- Sample Dissociation
- Sample Origin Taxonomy
- Sample Origin Type Taxonomy
- Sample Processing Taxonomy
- Sarcoidosis
- Scarlet Fever
- Seizure Disorder
- Serositis
- Severe
- Severity
- Severity Taxonomy
- Shoulder Dislocation
- Sign
- Sign or Symptom
- Simple Heterozygous
- Sinusitis
- Skin Basal Cell Carcinoma
- Skin Basosquamous Cell Carcinoma
- Skin Carcinoma
- Skin Cyst
- Somatic Variant
- Species Taxonomy
- Spina Bifida
- Spinal and Bulbar Muscular Atrophy, X-linked 1
- Staphylococcal Infection
- Stop Gained NMD Escaping
- Stroke
- Study Design
- Subdural Hematoma
- Symptom
- Syncope
- Synthetic Source
- Thalassemia
- Thoracic Trauma
- Thromboembolism
- Thyroid Gland Disorder
- Thyroiditis
- Tibia Fracture
- Tinea Versicolor
- Tooth Abscess
- Toxoplasmosis
- Transcriptomic Source
- Transient Ischemic Attack
- Trigeminal Nerve Disorder
- Tumor Grade Taxonomy
- Tumor Progression Taxonomy
- Two Vessel Coronary Disease
- Type 1 Diabtes Mellitus
- Type 2 Diabetes Mellitus
- Unit by Category
- Unit of Measure Taxonomy
- Unknown
- Ureteral Stenosis
- Uterine Corpus Leiomyoma
- Variant Origin
- Vertigo
- Viral Pericarditis
- Viral RNA Source
- Vitiligo
- Vulvovaginal Candidiasis
- Vulvovaginitis
- Zygosity
- Zygosity Taxonomy
5 Prime UTR Variantni back to ToC or Named Individual ToC
IRI: http://www.sequenceontology.org/browser/current_release/term/SO:0001623
- belongs to
- concept
- Genomic Feature Class c
Abdominal Painni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26682
- belongs to
- concept
- Disease, Disorder or Finding c
Abnormal Sampleni back to ToC or Named Individual ToC
IRI: http://www.ebi.ac.uk/efo/EFO_0009655
- belongs to
- concept
- Biosample Status c
Acquired Hypothyroidismni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C129644
- belongs to
- concept
- Disease, Disorder or Finding c
Acute Cystitisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26934
- belongs to
- concept
- Disease, Disorder or Finding c
Acute Myocardial Infarctionni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C35204
- belongs to
- concept
- Disease, Disorder or Finding c
Acute Pyelonephritisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C123215
- belongs to
- concept
- Disease, Disorder or Finding c
Acute Respiratory Failureni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C27043
- belongs to
- concept
- Disease, Disorder or Finding c
Acute Tonsillitisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C97142
- belongs to
- concept
- Disease, Disorder or Finding c
Adenocarcinomani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2852
- belongs to
- concept
- Disease, Disorder or Finding c
Alanine Aminotransferase Increasedni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26948
- belongs to
- concept
- Disease, Disorder or Finding c
Alcohol Dependanceni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C93040
- belongs to
- concept
- Disease, Disorder or Finding c
Allele Origin Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/alleleOriginTaxonomy
- belongs to
- concept scheme
Allergic Asthmani back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/389145006
- belongs to
- concept
- Disease, Disorder or Finding c
Allergic Rhinitisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C79532
- belongs to
- concept
- Disease, Disorder or Finding c
Alzheimer's Diseaseni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2866
- belongs to
- concept
- Disease, Disorder or Finding c
Amenorrheani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C61443
- belongs to
- concept
- Disease, Disorder or Finding c
Amenorrhea due to Pituitary Dysfunctionni back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/1144987003
- belongs to
- concept
- Disease, Disorder or Finding c
Amyotrophic Lateral Sclerosisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34373
- belongs to
- concept
- Disease, Disorder or Finding c
Anemiani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2869
- belongs to
- concept
- Disease, Disorder or Finding c
Angioedemani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C112175
- belongs to
- concept
- Disease, Disorder or Finding c
Ankle Fractureni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26989
- belongs to
- concept
- Disease, Disorder or Finding c
Ann Arbor Stage IV Mantle Cell Lymphomani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C8486
- belongs to
- concept
- Disease, Disorder or Finding c
Ann Arbor Stage IV T-Cell Non-Hodgkin Lymphomani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C8668
- belongs to
- concept
- Disease, Disorder or Finding c
Anorectal Fistulani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C60785
- belongs to
- concept
- Disease, Disorder or Finding c
Anorexiani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2875
- belongs to
- concept
- Disease, Disorder or Finding c
Anosmiani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C116369
- belongs to
- concept
- Disease, Disorder or Finding c
Anxiety Disorderni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2878
- belongs to
- concept
- Disease, Disorder or Finding c
Arrhythmiani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2881
- belongs to
- concept
- Disease, Disorder or Finding c
Arthritisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2883
- belongs to
- concept
- Disease, Disorder or Finding c
Asian (ethnic group)ni back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/315280000
- belongs to
- concept
- Group (social concept) c
Aspiration Pneumonitisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34932
- belongs to
- concept
- Disease, Disorder or Finding c
Assay Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/assayTaxonomy
- belongs to
- concept scheme
Asthmani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C28397
- belongs to
- concept
- Disease, Disorder or Finding c
Atopic Dermatitisni back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/24079001
- belongs to
- concept
- Disease, Disorder or Finding c
Atrial Fibrillationni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C50466
- belongs to
- concept
- Disease, Disorder or Finding c
Atrial Flutterni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C51224
- belongs to
- concept
- Disease, Disorder or Finding c
Autoimmune Diseaseni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2889
- belongs to
- concept
- Disease, Disorder or Finding c
Autoimmune Nervous System Disorderni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C99383
- belongs to
- concept
- Disease, Disorder or Finding c
Autoimmune Pancytopeniani back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/183005
- belongs to
- concept
- Disease, Disorder or Finding c
Bacterial Pneumoniani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26704
- belongs to
- concept
- Disease, Disorder or Finding c
Baker Cystni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34935
- belongs to
- concept
- Disease, Disorder or Finding c
Basal Cell Carcinomani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C156767
- belongs to
- concept
- Disease, Disorder or Finding c
Bell's Palsyni back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/193093009
- belongs to
- concept
- Disease, Disorder or Finding c
Biosample Status Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/biosampleStatusTaxonomy
- belongs to
- concept scheme
Bipolar Disorderni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34423
- belongs to
- concept
- Disease, Disorder or Finding c
Black (ethnic group)ni back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/315240009
- belongs to
- concept
- Group (social concept) c
Blepharititsni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C112183
- belongs to
- concept
- Disease, Disorder or Finding c
Blepharospasmni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C118723
- belongs to
- concept
- Disease, Disorder or Finding c
Borderlineni back to ToC or Named Individual ToC
IRI: https://hpo.jax.org/browse/term/HP:0012827
Bowel Obstructionni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C9175
- belongs to
- concept
- Disease, Disorder or Finding c
Bradycardiani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C37920
- belongs to
- concept
- Disease, Disorder or Finding c
Breast Carcinomani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C4872
- belongs to
- concept
- Disease, Disorder or Finding c
Brown-Sequard Syndromeni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C84601
- belongs to
- concept
- Disease, Disorder or Finding c
Cancer Cell Line Sampleni back to ToC or Named Individual ToC
IRI: http://www.ebi.ac.uk/efo/EFO_0030035
- belongs to
- concept
- Biosample Status c
Cancer TNM Finding Categoryni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C48698
- belongs to
- concept
- Pathological TNM Finding c
Carcinomani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2916
- belongs to
- concept
- Disease, Disorder or Finding c
Cardiovascular Disorderni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2931
- belongs to
- concept
- Disease, Disorder or Finding c
Cardiovascular Systemni back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/UATC/C
- belongs to
- concept
- pharmacologic substance c
Case Control Designni back to ToC or Named Individual ToC
IRI: http://www.ebi.ac.uk/efo/EFO_0001427
- belongs to
- concept
- Biosample Status c
Caucasianni back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/14045001
- belongs to
- concept
- Group (social concept) c
Cavernous Hemangiomani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3086
- belongs to
- concept
- Disease, Disorder or Finding c
Celiac Diseaseni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26714
- belongs to
- concept
- Disease, Disorder or Finding c
Central Nervous System Degenerative Disorderni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C4802
- belongs to
- concept
- Disease, Disorder or Finding c
Chicken Poxni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C97132
- belongs to
- concept
- Disease, Disorder or Finding c
Chronic Lung Disorderni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C98541
- belongs to
- concept
- Disease, Disorder or Finding c
Clinical Modifierni back to ToC or Named Individual ToC
IRI: https://hpo.jax.org/browse/term/HP:0012823
- belongs to
- concept
- Phenotypic Feature Modifier c
Closed Dislocation of Shoulderni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34486
- belongs to
- concept
- Disease, Disorder or Finding c
Closed Fracture of Ribni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C35350
- belongs to
- concept
- Disease, Disorder or Finding c
Closed Fracture of Tibiani back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/447139008
- belongs to
- concept
- Disease, Disorder or Finding c
Cognitive Imparimentni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C116921
- belongs to
- concept
- Disease, Disorder or Finding c
Cohort Data Type Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/cohortDataTypeTaxonomy
- belongs to
- concept scheme
Cohort Design Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/cohortDesignTaxonomy
- belongs to
- concept scheme
Cohort Sex Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/cohortSexTaxonomy
- belongs to
- concept scheme
Cold Soreni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34695
- belongs to
- concept
- Disease, Disorder or Finding c
Colon Adenocarcinomani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C4349
- belongs to
- concept
- Disease, Disorder or Finding c
Common Bile Duct Stoneni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34499
- belongs to
- concept
- Disease, Disorder or Finding c
Complete Genomic Deletionni back to ToC or Named Individual ToC
IRI: http://www.ebi.ac.uk/efo/EFO_0030069
- belongs to
- concept
- Copy Number Assessment c
Compouns Heterozygousni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/GENO_0000402
Congenital Adrenal Hyperplasiani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34360
- belongs to
- concept
- Disease, Disorder or Finding c
Copy Number Assessmentni back to ToC or Named Individual ToC
IRI: http://www.ebi.ac.uk/efo/EFO_0030063
- belongs to
- concept
- Copy Number Assessment c
Copy Number Assessment Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/copyNumberAssessmentTaxonomy
- belongs to
- concept scheme
Copy Number Gainni back to ToC or Named Individual ToC
IRI: http://www.ebi.ac.uk/efo/EFO_0030070
- belongs to
- concept
- Copy Number Assessment c
Copy Number Lossni back to ToC or Named Individual ToC
IRI: http://www.ebi.ac.uk/efo/EFO_0030067
- belongs to
- concept
- Copy Number Assessment c
Coronary Artery Bypass Graftni back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/232717009
- belongs to
- concept
- surgical type c
COVID-19 Infectionni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C171133
- belongs to
- concept
- Disease, Disorder or Finding c
Cranial Nerve VII Palsyni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26769
- belongs to
- concept
- Disease, Disorder or Finding c
Cutaneous Melanomani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3510
- belongs to
- concept
- Disease, Disorder or Finding c
Cystni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2978
- belongs to
- concept
- Disease, Disorder or Finding c
De-Novo Variantni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/SO_0001781
- belongs to
- concept
- Allele Origin c
Deep Vein Thrombosisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C49343
- belongs to
- concept
- Disease, Disorder or Finding c
Dementiani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C4786
- belongs to
- concept
- Disease, Disorder or Finding c
Depressionni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2982
- belongs to
- concept
- Disease, Disorder or Finding c
Diabetes Mellitusni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2985
- belongs to
- concept
- Disease, Disorder or Finding c
Diabetic Ketoacidosisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C50530
- belongs to
- concept
- Disease, Disorder or Finding c
Diagnostic Marker Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/diagnosticMarkerTaxonomy
- belongs to
- concept scheme
Disease Grade Qualifierni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C28076
- belongs to
- concept
- Tumor Grade c
Disease or Disorderni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2991
- belongs to
- concept
- Disease, Disorder or Finding c
Disease or Disorder (MONDO)ni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/MONDO_0000001
- belongs to
- concept
- PhenoClinic Category c
Disease Stage Qualifierni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C28108
- belongs to
- concept
- Pathological Stage c
Disease Stage Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/diseaseStageTaxonomy
- belongs to
- concept scheme
Disease, Disorder or Finding Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/phenoclinical/schema/diseaseDisorderFindingTaxonomy
- belongs to
- concept scheme
DNA Sequencerni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/OBI_0400103
- belongs to
- concept
- Platform Model c
Dog Biteni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C122419
- belongs to
- concept
- Disease, Disorder or Finding c
Drug Hypersensitivity Syndromeni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C112208
- belongs to
- concept
- Disease, Disorder or Finding c
Dyslipidemiani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C80385
- belongs to
- concept
- Disease, Disorder or Finding c
Eczemani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3001
- belongs to
- concept
- Disease, Disorder or Finding c
Endometriosisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3014
- belongs to
- concept
- Disease, Disorder or Finding c
Epidermal Inclusion Cystni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3134
- belongs to
- concept
- Disease, Disorder or Finding c
Essential Hypertensionni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3478
- belongs to
- concept
- Disease, Disorder or Finding c
Ethnic Groupni back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/372148003
- belongs to
- concept
- Group (social concept) c
Experimental Evidenceni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/ECO_0000006
- belongs to
- concept
- PhenoClinic Evidence c
Extrapyramidal Disorderni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C79593
- belongs to
- concept
- Disease, Disorder or Finding c
Favismni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34607
- belongs to
- concept
- Disease, Disorder or Finding c
Femaleni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C16576
- belongs to
- concept
- Cohort Sex c
Femur Fractureni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26774
- belongs to
- concept
- Disease, Disorder or Finding c
Fever Chillsni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26921
- belongs to
- concept
- Disease, Disorder or Finding c
Fibromani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3041
- belongs to
- concept
- Disease, Disorder or Finding c
Fibromyalgiani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C87497
- belongs to
- concept
- Disease, Disorder or Finding c
Findingni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3367
- belongs to
- concept
- Disease, Disorder or Finding c
Flu-Like Symptomsni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C78302
- belongs to
- concept
- Disease, Disorder or Finding c
Fungal Infectionni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3245
- belongs to
- concept
- Disease, Disorder or Finding c
Gastritisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26780
- belongs to
- concept
- Disease, Disorder or Finding c
Gastroenterititsni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34632
- belongs to
- concept
- Disease, Disorder or Finding c
Gastroesophageal Refluxni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C92560
- belongs to
- concept
- Disease, Disorder or Finding c
Gastroplastyni back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/9429009
- belongs to
- concept
- surgical type c
Generalized Epilepsyni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3021
- belongs to
- concept
- Disease, Disorder or Finding c
Genomic Feature Class Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/genomicFeatureClassTaxonomy
- belongs to
- concept scheme
Genomic Sourceni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/GENEPIO_0001966
- belongs to
- concept
- Library Source c
Geographic Location Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/geographicLocationTaxonomy
- belongs to
- concept scheme
Germline Variantni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/SO_0001778
- belongs to
- concept
- Allele Origin c
Gianmaria Silvelloni back to ToC or Named Individual ToC
IRI: https://orcid.org/0000-0003-4970-4554
- belongs to
- Person c
Glaucomani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26782
- belongs to
- concept
- Disease, Disorder or Finding c
Group Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/phenoclinical/schema/groupTaxonomy
- belongs to
- concept scheme
Hashimoto Thyroiditisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C27191
- belongs to
- concept
- Disease, Disorder or Finding c
Head and Neck Findingni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C36282
- belongs to
- concept
- Disease, Disorder or Finding c
Head Traumani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34660
- belongs to
- concept
- Disease, Disorder or Finding c
Headacheni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34661
- belongs to
- concept
- Disease, Disorder or Finding c
Heart Failureni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C50577
- belongs to
- concept
- Disease, Disorder or Finding c
Hemizygousni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/GENO_0000134
Hemizygous Insertion-Linkedni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/GENO_0000606
Hemizygous X-Linkedni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/GENO_0000604
Hemizygous Y-Linkedni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/GENO_0000605
Hepatic Toxicityni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C185645
- belongs to
- concept
- Disease, Disorder or Finding c
Hepatitisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3095
- belongs to
- concept
- Disease, Disorder or Finding c
Hepatitis A Infectionni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3096
- belongs to
- concept
- Disease, Disorder or Finding c
Hepatitis B, Purified Antigenni back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/UATC/J07BC01
- belongs to
- concept
- pharmacologic substance c
Hereditary Factor XI Deficiencyni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C84705
- belongs to
- concept
- Disease, Disorder or Finding c
Hereditary Hemolytic Anemiani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34379
- belongs to
- concept
- Disease, Disorder or Finding c
Herniani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34685
- belongs to
- concept
- Disease, Disorder or Finding c
Herpes Simplex Keratitisni back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/9389005
- belongs to
- concept
- Disease, Disorder or Finding c
Herpes Simplex Virus Infectionni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C155871
- belongs to
- concept
- Disease, Disorder or Finding c
Herpes Zosterni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C71079
- belongs to
- concept
- Disease, Disorder or Finding c
Heterozygousni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/GENO_0000391
High Grade Cervical Squamous Intraepithelial Neoplasiani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C40197
- belongs to
- concept
- Disease, Disorder or Finding c
High-Level Copy Number Gainni back to ToC or Named Individual ToC
IRI: http://www.ebi.ac.uk/efo/EFO_0030072
- belongs to
- concept
- Copy Number Assessment c
High-Level Copy Number Lossni back to ToC or Named Individual ToC
IRI: http://www.ebi.ac.uk/efo/EFO_0020073
- belongs to
- concept
- Copy Number Assessment c
Hispanicni back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/414408004
- belongs to
- concept
- Group (social concept) c
Homozygousni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/GENO_0000136
Humanni back to ToC or Named Individual ToC
IRI: https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=9606
Hypersensitivityni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3114
- belongs to
- concept
- Disease, Disorder or Finding c
Hypertensionni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3117
- belongs to
- concept
- Disease, Disorder or Finding c
Hypertensive Crisisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3122
- belongs to
- concept
- Disease, Disorder or Finding c
Hyperthyroidismni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3123
- belongs to
- concept
- Disease, Disorder or Finding c
Hypothyroidismni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26800
- belongs to
- concept
- Disease, Disorder or Finding c
Infectious Mononucleosisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34726
- belongs to
- concept
- Disease, Disorder or Finding c
Inferential Evidenceni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/ECO_0000361
- belongs to
- concept
- PhenoClinic Evidence c
Inguinal Herniani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34690
- belongs to
- concept
- Disease, Disorder or Finding c
Injuryni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3671
- belongs to
- concept
- Disease, Disorder or Finding c
Interstitial Pneumoniani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C27006
- belongs to
- concept
- Disease, Disorder or Finding c
Intestinal Pseudo-Obstructionni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34733
- belongs to
- concept
- Disease, Disorder or Finding c
Iridocyclitisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34736
- belongs to
- concept
- Disease, Disorder or Finding c
Iron-Deficiency Anemiani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C84484
- belongs to
- concept
- Disease, Disorder or Finding c
Keratitisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26805
- belongs to
- Disease, Disorder or Finding c
Keratoconusni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26806
- belongs to
- concept
- Disease, Disorder or Finding c
Klippel-Feil Syndromeni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C98967
- belongs to
- concept
- Disease, Disorder or Finding c
Langerhans Cell Histiocytosisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3107
- belongs to
- concept
- Disease, Disorder or Finding c
Laura Menottini back to ToC or Named Individual ToC
IRI: https://orcid.org/0000-0002-0676-682X
- belongs to
- Person c
Leukopeniani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26816
- belongs to
- concept
- Disease, Disorder or Finding c
Library Sourceni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/GENEPIO_0001965
- belongs to
- concept
- Library Source c
Library Source Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/librarySourceTaxonomy
- belongs to
- concept scheme
Long QT Syndromeni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34786
- belongs to
- concept
- Disease, Disorder or Finding c
Low-Level Copy Number Gainni back to ToC or Named Individual ToC
IRI: http://www.ebi.ac.uk/efo/EFO_0030071
- belongs to
- concept
- Copy Number Assessment c
Low-Level Copy Number Lossni back to ToC or Named Individual ToC
IRI: http://www.ebi.ac.uk/efo/EFO_0030068
- belongs to
- concept
- Copy Number Assessment c
Lumbar Herniani back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/23344004
- belongs to
- concept
- Disease, Disorder or Finding c
Lumbar Puncture Headacheni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C50643
- belongs to
- concept
- Disease, Disorder or Finding c
Lymphopeniani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26823
- belongs to
- concept
- Disease, Disorder or Finding c
Maleni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C20197
- belongs to
- concept
- Cohort Sex c
Malignant Colon Neoplasmni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C9242
- belongs to
- concept
- Disease, Disorder or Finding c
Malignant Neoplasmni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C9305
- belongs to
- concept
- Disease, Disorder or Finding c
Malignant Skin Neoplasmni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2920
- belongs to
- concept
- Disease, Disorder or Finding c
Malignant Thyroid Gland Neoplasmni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C7510
- belongs to
- concept
- Disease, Disorder or Finding c
Material Entityni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/BFO_0000040
- belongs to
- concept
- Sample Origin Type c
Maternal Variantni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/SO_0001775
- belongs to
- concept
- Allele Origin c
Melanocytic Nevusni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C7570
- belongs to
- concept
- Disease, Disorder or Finding c
Metagenomic Sourceni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/GENEPIO_0001967
- belongs to
- concept
- Library Source c
Metastasis Sampleni back to ToC or Named Individual ToC
IRI: http://www.ebi.ac.uk/efo/EFO_0010941
- belongs to
- concept
- Biosample Status c
Metatranscriptomic Sourceni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/GENEPIO_0001968
- belongs to
- concept
- Library Source c
Migraineni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C89715
- belongs to
- concept
- Disease, Disorder or Finding c
Migraine With Aurani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C117005
- belongs to
- concept
- Disease, Disorder or Finding c
Mildni back to ToC or Named Individual ToC
IRI: https://hpo.jax.org/browse/term/HP:0012825
Missense Variantni back to ToC or Named Individual ToC
IRI: http://www.sequenceontology.org/browser/current_release/term/SO:0001583
- belongs to
- concept
- Molecular Effect c
Moderateni back to ToC or Named Individual ToC
IRI: https://hpo.jax.org/browse/term/HP:0012826
Molecular Effect Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/molecularEffectTaxonomy
- belongs to
- concept scheme
Molluscum Contagiosum Virusni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C112357
- belongs to
- concept
- Disease, Disorder or Finding c
Monoclonal B_Cell Lymphocytosisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C80310
- belongs to
- concept
- Disease, Disorder or Finding c
Monoclonal Gammopathyni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C35548
- belongs to
- concept
- Disease, Disorder or Finding c
Mood Disorderni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C92200
- belongs to
- concept
- Disease, Disorder or Finding c
Motor Neuron Diseaseni back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/37340000
- belongs to
- concept
- Disease, Disorder or Finding c
Multiple Sclerosisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3243
- belongs to
- concept
- Disease, Disorder or Finding c
Mumpsni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C29888
- belongs to
- concept
- Disease, Disorder or Finding c
Myasthenia Gravisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C60989
- belongs to
- concept
- Disease, Disorder or Finding c
Myeloproliferative Neoplasmni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C4345
- belongs to
- concept
- Disease, Disorder or Finding c
Neck Traumani back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/90460009
- belongs to
- concept
- Disease, Disorder or Finding c
Neonatal Hearing Impairmentni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C50667
- belongs to
- concept
- Disease, Disorder or Finding c
Neoplasmni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3262
- belongs to
- concept
- Disease, Disorder or Finding c
Neoplasm by Special Categoryni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C7062
- belongs to
- concept
- Tumor Progression c
Neoplastic Sampleni back to ToC or Named Individual ToC
IRI: http://www.ebi.ac.uk/efo/EFO_0009656
- belongs to
- concept
- Biosample Status c
Neurofibromatosisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C6727
- belongs to
- concept
- Disease, Disorder or Finding c
Neutropenic Disorderni back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/303011007
- belongs to
- concept
- Disease, Disorder or Finding c
Non-Neoplastic Central Nervous System Disorderni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C27586
- belongs to
- concept
- Disease, Disorder or Finding c
Nullizygousni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/GENO_0000978
Obesityni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3283
- belongs to
- concept
- Disease, Disorder or Finding c
Obstructive Sleep Apnea Syndromeni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C27168
- belongs to
- concept
- Disease, Disorder or Finding c
Obtention Procedure Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/obtentionProcedureTaxonomy
- belongs to
- concept scheme
Opioid Use Disorderni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C157942
- belongs to
- concept
- Disease, Disorder or Finding c
Optic Neuritisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C84950
- belongs to
- concept
- Disease, Disorder or Finding c
Osteoporosisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3298
- belongs to
- concept
- Disease, Disorder or Finding c
Other Library Sourceni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/GENEPIO_0001969
- belongs to
- concept
- Library Source c
Palmar Fibromatosisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3469
- belongs to
- concept
- Disease, Disorder or Finding c
Pancytopeniani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34889
- belongs to
- concept
- Disease, Disorder or Finding c
Parkinson's Diseaseni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26845
- belongs to
- concept
- Disease, Disorder or Finding c
Paternal Variantni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/SO_0001776
- belongs to
- concept
- Allele Origin c
Pathological Stage Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/pathologicalStageTaxonomy
- belongs to
- concept scheme
Pathological TNM Finding Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/pathologicalTNMFindingTaxonomy
- belongs to
- concept scheme
Pedigree Specific Variantni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/SO_0001779
- belongs to
- concept
- Allele Origin c
Penicillin Allergyni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34911
- belongs to
- concept
- Disease, Disorder or Finding c
Personality Disorderni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34922
- belongs to
- concept
- Disease, Disorder or Finding c
Phenoclinic Category Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/phenoClinicCategoryTaxonomy
- belongs to
- concept scheme
Phenoclinic Evidence Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/phenoclinicEvidenceTaxonomy
- belongs to
- concept scheme
Phenotypic Abnormalityni back to ToC or Named Individual ToC
IRI: https://hpo.jax.org/browse/term/HP:0000118
- belongs to
- concept
- PhenoClinic Category c
Phenotypic Feature Modifier Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/phenotypicFeatureModifierTaxonomy
- belongs to
- concept scheme
Phenotypic Feature Type Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/phenotypicFeatureTypeTaxonomy
- belongs to
- concept scheme
Pituitary Neuroendocrine Tumorni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3329
- belongs to
- concept
- Disease, Disorder or Finding c
Platform Model Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/platformModelTaxonomy
- belongs to
- concept scheme
Pleural Effusionni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3331
- belongs to
- concept
- Disease, Disorder or Finding c
Pneumococcal Pneumoniani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C157959
- belongs to
- concept
- Disease, Disorder or Finding c
Pneumoniani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3333
- belongs to
- concept
- Disease, Disorder or Finding c
Pneumonia Caused By SARS-CoV-2ni back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/882784691000119100
- belongs to
- concept
- Disease, Disorder or Finding c
Population Specific Variantni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/SO_0001780
- belongs to
- concept
- Allele Origin c
Post-Herpetic Neuralgiani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C117084
- belongs to
- concept
- Disease, Disorder or Finding c
Primary Neoplasmni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C8509
- belongs to
- concept
- Disease, Disorder or Finding c
Primary Progressive Aphasiani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C85024
- belongs to
- concept
- Disease, Disorder or Finding c
Primary Tumor Sampleni back to ToC or Named Individual ToC
IRI: http://www.ebi.ac.uk/efo/EFO_0010942
- belongs to
- concept
- Biosample Status c
Profoundni back to ToC or Named Individual ToC
IRI: https://hpo.jax.org/browse/term/HP:0012829
Psoriasisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3346
- belongs to
- concept
- Disease, Disorder or Finding c
Psoriatic Arthritisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C61277
- belongs to
- concept
- Disease, Disorder or Finding c
Psychiatric Disorderni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2893
- belongs to
- concept
- Disease, Disorder or Finding c
Pulmonary Embolismni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C50713
- belongs to
- concept
- Disease, Disorder or Finding c
Pulmonary Sarcoidosisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34997
- belongs to
- concept
- Disease, Disorder or Finding c
Pulmonary Thromboembolismni back to ToC or Named Individual ToC
IRI: http://purl.bioontology.org/ontology/SNOMEDCT/233935004
- belongs to
- concept
- Disease, Disorder or Finding c
Radius Fractureni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C99039
- belongs to
- concept
- Disease, Disorder or Finding c
Recurrent Tumor Sampleni back to ToC or Named Individual ToC
IRI: http://www.ebi.ac.uk/efo/EFO_0010943
- belongs to
- concept
- Biosample Status c
Reference Sampleni back to ToC or Named Individual ToC
IRI: http://www.ebi.ac.uk/efo/EFO_0009654
- belongs to
- concept
- Biosample Status c
Regional Base Ploidyni back to ToC or Named Individual ToC
IRI: http://www.ebi.ac.uk/efo/EFO_0030064
- belongs to
- concept
- Copy Number Assessment c
Renal Colicni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C78593
- belongs to
- concept
- Disease, Disorder or Finding c
Retinal Detachmentni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26874
- belongs to
- concept
- Disease, Disorder or Finding c
Rheumatoid Arthritisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2884
- belongs to
- concept
- Disease, Disorder or Finding c
Sample Dissociationni back to ToC or Named Individual ToC
IRI: http://www.ebi.ac.uk/efo/EFO_0009091
- belongs to
- concept
- Sample Processing c
Sample Origin Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/sampleOriginDetailTaxonomy
- belongs to
- concept scheme
Sample Origin Type Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/sampleOriginTypeTaxonomy
- belongs to
- concept scheme
Sample Processing Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/sampleProcessingTaxonomy
- belongs to
- concept scheme
Sarcoidosisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C34995
- belongs to
- concept
- Disease, Disorder or Finding c
Scarlet Feverni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C94575
- belongs to
- concept
- Disease, Disorder or Finding c
Seizure Disorderni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3020
- belongs to
- concept
- Disease, Disorder or Finding c
Serositisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C70428
- belongs to
- concept
- Disease, Disorder or Finding c
Severeni back to ToC or Named Individual ToC
IRI: https://hpo.jax.org/browse/term/HP:0012828
Severityni back to ToC or Named Individual ToC
IRI: https://hpo.jax.org/browse/term/HP:0012824
Severity Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/severityTaxonomy
- belongs to
- concept scheme
Shoulder Dislocationni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C35020
- belongs to
- concept
- Disease, Disorder or Finding c
Signni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C53458
- belongs to
- concept
- Disease, Disorder or Finding c
Sign or Symptomni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C100104
- belongs to
- concept
- Disease, Disorder or Finding c
Simple Heterozygousni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/GENO_0000458
Sinusitisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C35024
- belongs to
- concept
- Disease, Disorder or Finding c
Skin Basal Cell Carcinomani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2921
- belongs to
- concept
- Disease, Disorder or Finding c
Skin Basosquamous Cell Carcinomani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2922
- belongs to
- concept
- Disease, Disorder or Finding c
Skin Carcinomani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C4914
- belongs to
- concept
- Disease, Disorder or Finding c
Skin Cystni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3884
- belongs to
- concept
- Disease, Disorder or Finding c
Somatic Variantni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/SO_0001777
- belongs to
- concept
- Allele Origin c
Species Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/speciesTaxonomy
- belongs to
- concept scheme
Spina Bifidani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C101214
- belongs to
- concept
- Disease, Disorder or Finding c
Spinal and Bulbar Muscular Atrophy, X-linked 1ni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C85233
- belongs to
- concept
- Disease, Disorder or Finding c
Staphylococcal Infectionni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C35038
- belongs to
- concept
- Disease, Disorder or Finding c
Stop Gained NMD Escapingni back to ToC or Named Individual ToC
IRI: http://www.sequenceontology.org/browser/current_release/term/SO:0002322
- belongs to
- concept
- Molecular Effect c
Strokeni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3390
- belongs to
- concept
- Disease, Disorder or Finding c
Study Designni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/OBI_0500000
- belongs to
- concept
- Cohort Design c
Subdural Hematomani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C116585
- belongs to
- concept
- Disease, Disorder or Finding c
Symptomni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C4876
- belongs to
- concept
- Disease, Disorder or Finding c
Syncopeni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C35053
- belongs to
- concept
- Disease, Disorder or Finding c
Synthetic Sourceni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/GENEPIO_0001970
- belongs to
- concept
- Library Source c
Thalassemiani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C35069
- belongs to
- concept
- Disease, Disorder or Finding c
Thoracic Traumani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C171204
- belongs to
- concept
- Disease, Disorder or Finding c
Thromboembolismni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C28195
- belongs to
- concept
- Disease, Disorder or Finding c
Thyroid Gland Disorderni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26893
- belongs to
- concept
- Disease, Disorder or Finding c
Thyroiditisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26894
- belongs to
- concept
- Disease, Disorder or Finding c
Tibia Fractureni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C99083
- belongs to
- concept
- Disease, Disorder or Finding c
Tinea Versicolorni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C112833
- belongs to
- concept
- Disease, Disorder or Finding c
Tooth Abscessni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C35475
- belongs to
- concept
- Disease, Disorder or Finding c
Toxoplasmosisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3418
- belongs to
- concept
- Disease, Disorder or Finding c
Transcriptomic Sourceni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/GENEPIO_0001971
- belongs to
- concept
- Library Source c
Transient Ischemic Attackni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C50781
- belongs to
- concept
- Disease, Disorder or Finding c
Trigeminal Nerve Disorderni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26952
- belongs to
- concept
- Disease, Disorder or Finding c
Tumor Grade Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/tumorGradeTaxonomy
- belongs to
- concept scheme
Tumor Progression Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/tumorProgressionTaxonomy
- belongs to
- concept scheme
Two Vessel Coronary Diseaseni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C100023
- belongs to
- concept
- Disease, Disorder or Finding c
Type 1 Diabtes Mellitusni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2986
- belongs to
- concept
- Disease, Disorder or Finding c
Type 2 Diabetes Mellitusni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26747
- belongs to
- concept
- Disease, Disorder or Finding c
Unit by Categoryni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C42568
- belongs to
- concept
- Unit Of Measure c
Unit of Measure Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/uomTaxonomy
- belongs to
- concept scheme
Unknownni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C17998
- belongs to
- concept
- Cohort Sex c
Ureteral Stenosisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C115958
- belongs to
- concept
- Disease, Disorder or Finding c
Uterine Corpus Leiomyomani back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C3434
- belongs to
- concept
- Disease, Disorder or Finding c
Variant Originni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/SO_0001762
- belongs to
- concept
- Allele Origin c
Vertigoni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C38057
- belongs to
- concept
- Disease, Disorder or Finding c
Viral Pericarditisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C128405
- belongs to
- concept
- Disease, Disorder or Finding c
Viral RNA Sourceni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/GENEPIO_0001972
- belongs to
- concept
- Library Source c
Vitiligoni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C26915
- belongs to
- concept
- Disease, Disorder or Finding c
Vulvovaginal Candidiasisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C2914
- belongs to
- concept
- Disease, Disorder or Finding c
Vulvovaginitisni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/NCIT_C35131
- belongs to
- concept
- Disease, Disorder or Finding c
Zygosityni back to ToC or Named Individual ToC
IRI: http://purl.obolibrary.org/obo/GENO_0000133
Zygosity Taxonomyni back to ToC or Named Individual ToC
IRI: https://w3id.org/hereditary/ontology/genomics/schema/zigosityTaxonomy
- belongs to
- concept scheme
Legend back to ToC
op: Object Properties
dp: Data Properties
ni: Named Individuals


